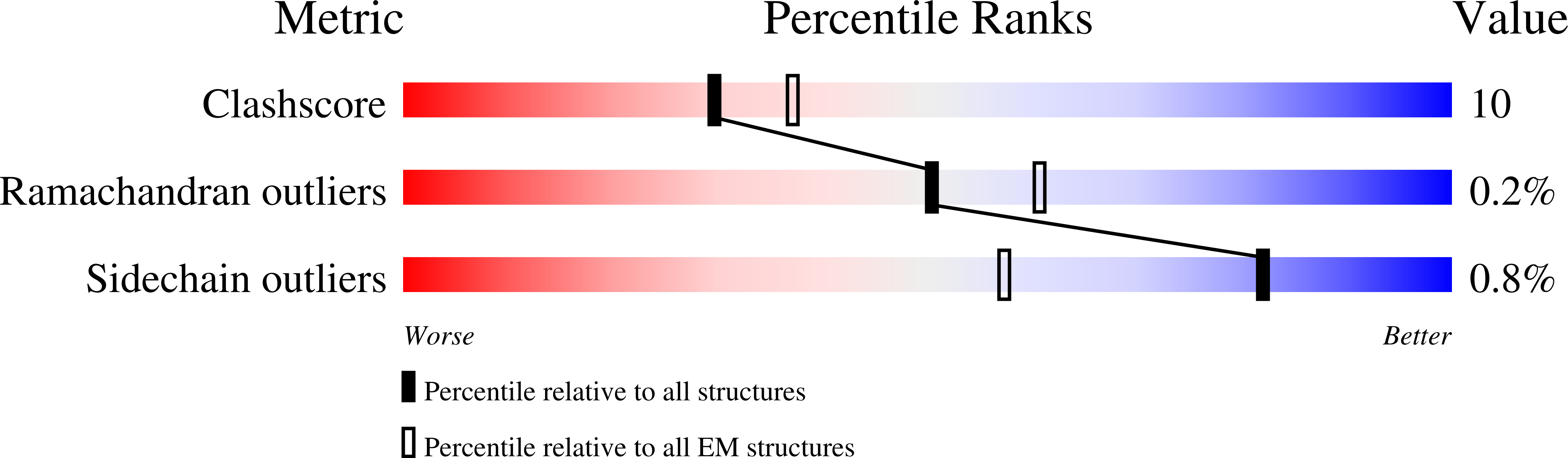Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Twomey, E.C., Ji, Z., Wales, T.E., Bodnar, N.O., Ficarro, S.B., Marto, J.A., Engen, J.R., Rapoport, T.A.(2019) Science 365
- PubMed: 31249135
- DOI: https://doi.org/10.1126/science.aax1033
- Primary Citation of Related Structures:
6OA9, 6OAA, 6OAB - PubMed Abstract:
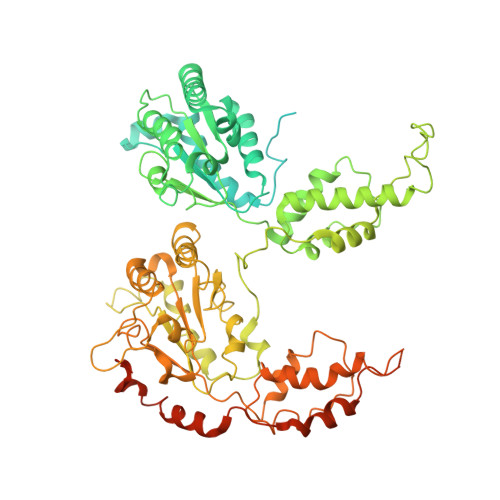
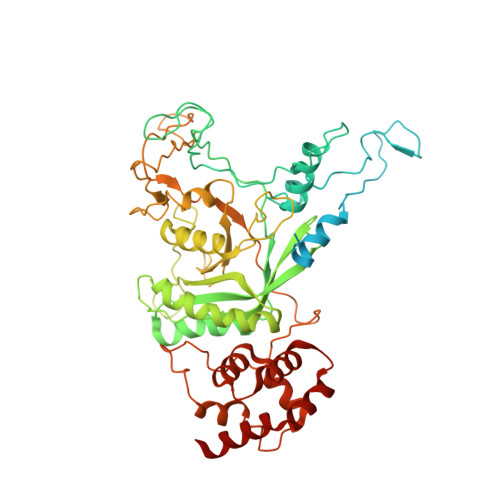
The Cdc48 adenosine triphosphatase (ATPase) (p97 or valosin-containing protein in mammals) and its cofactor Ufd1/Npl4 extract polyubiquitinated proteins from membranes or macromolecular complexes for subsequent degradation by the proteasome. How Cdc48 processes its diverse and often well-folded substrates is unclear. Here, we report cryo-electron microscopy structures of the Cdc48 ATPase in complex with Ufd1/Npl4 and polyubiquitinated substrate. The structures show that the Cdc48 complex initiates substrate processing by unfolding a ubiquitin molecule. The unfolded ubiquitin molecule binds to Npl4 and projects its N-terminal segment through both hexameric ATPase rings. Pore loops of the second ring form a staircase that acts as a conveyer belt to move the polypeptide through the central pore. Inducing the unfolding of ubiquitin allows the Cdc48 ATPase complex to process a broad range of substrates.
Organizational Affiliation:
Department of Cell Biology, Harvard Medical School, and Howard Hughes Medical Institute, 240 Longwood Avenue, Boston, MA 02115, USA.