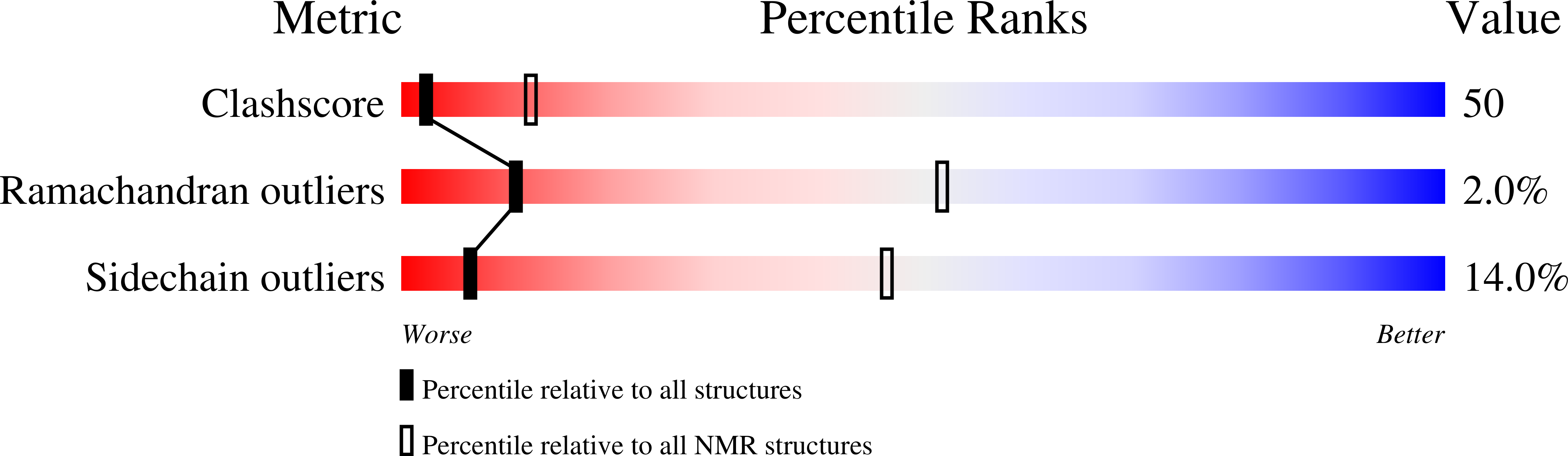Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Pinheiro-Aguiar, R., do Amaral, V.S.G., Pereira, I.B., Kurtenbach, E., Almeida, F.C.L.(2020) Proteins 88: 242-246
- PubMed: 31294889
- DOI: https://doi.org/10.1002/prot.25783
- Primary Citation of Related Structures:
6NOM - PubMed Abstract:
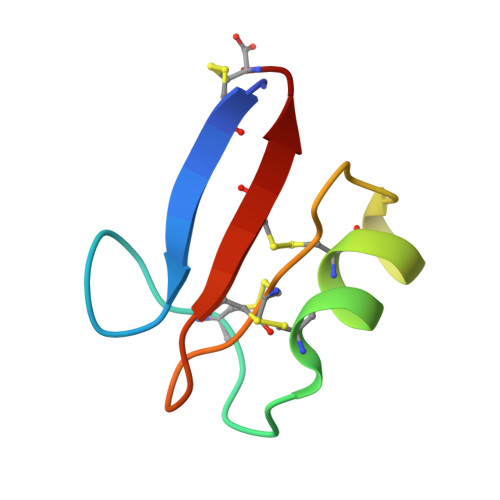
Pisum sativum defensin 2 (Psd2) is a small (4.7 kDa) antifungal peptide whose structure is held together by four conserved disulfide bridges. Psd2 shares the cysteine-stabilized alpha-beta (CSαβ) fold, which lacks a regular hydrophobic core. All hydrophobic residues are exposed to the surface, except for leucine 6. They are clustered in the surface formed by two loops, between β1 and α-helix and β2 and β3 sheets. The observation of surface hydrophobic clusters reveals a remarkable evolution of the CSαβ fold to expose and reorganize hydrophobic residues, which facilitates creating versatile binding sites.
Organizational Affiliation:
Instituto de Bioquímica Médica Leopoldo de Meis, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brasil.