Noncovalent structure of SENP1 in complex with SUMO2.
Ambaye, N.D.(2019) Acta Crystallogr F Struct Biol Commun 75: 332-339
- PubMed: 31045562
- DOI: https://doi.org/10.1107/S2053230X19004266
- Primary Citation of Related Structures:
6NNQ - PubMed Abstract:
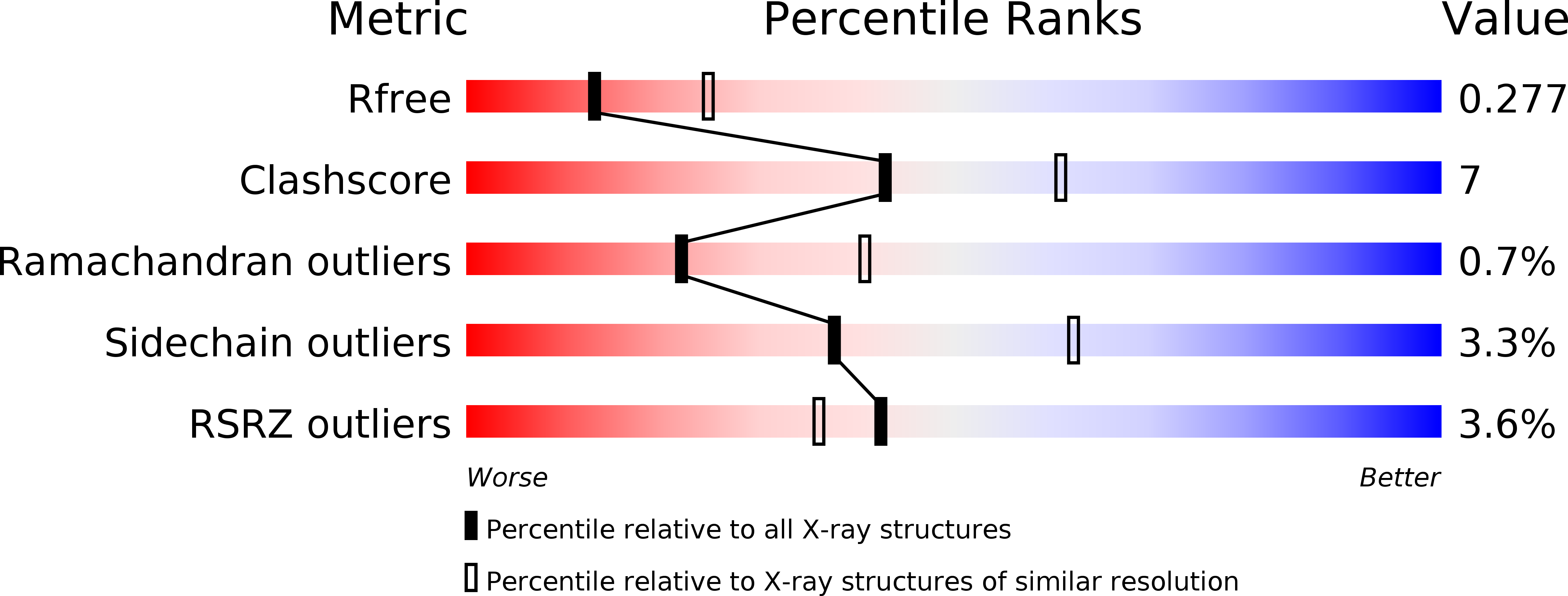
SUMOylation is a post-translational modification in which a small ubiquitin-like molecule (SUMO) is appended to substrate proteins and is known to influence myriads of biological processes. A delicate interplay between several families of SUMOylation proteins and their substrates ensures the proper level of SUMOylation required for normal cell function. Among the SUMO proteins, SUMO2 is known to form mono-SUMOylated proteins and engage in poly-SUMO chain formation, while sentrin-specific protease 1 (SENP1) is a key enzyme in regulating both events. Determination of the SENP1-SUMO2 interaction is therefore necessary to better understand SUMOylation. In this regard, the current paper reports the noncovalent structure of SENP1 in complex with SUMO2, which was refined to a resolution of 2.62 Å with R and R free values of 22.92% and 27.66%, respectively. The structure shows that SENP1-SUMO2 complex formation is driven largely by polar interactions and limited hydrophobic contacts. The essential C-terminal motif (QQTGG) of SUMO2 is stabilized by a number of specific bonding interactions that enable it to protrude into the catalytic triad of SENP1 and provide the arrangement necessary for maturation of SUMO and deSUMOylation activity. Overall, the structure shows a number of structural details that pinpoint the basis of SENP1-SUMO2 complex formation.
Organizational Affiliation:
Department of Immune-Oncology, Beckman Research Institute, City of Hope National Medical Center, 1500 East Duarte Road, Duarte, CA 91010, USA.