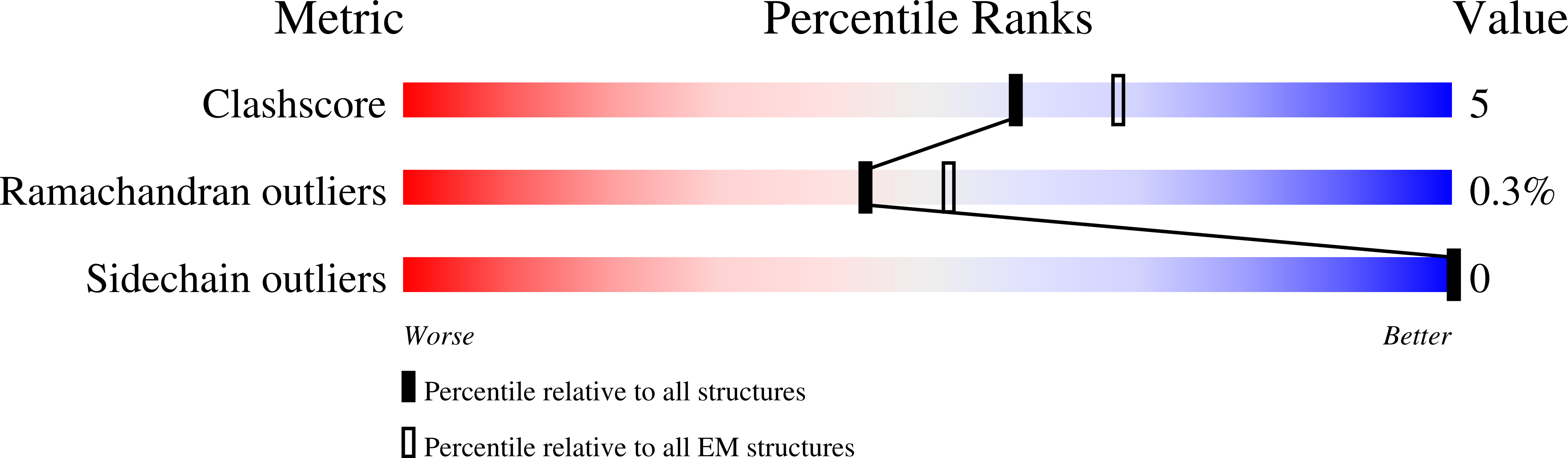
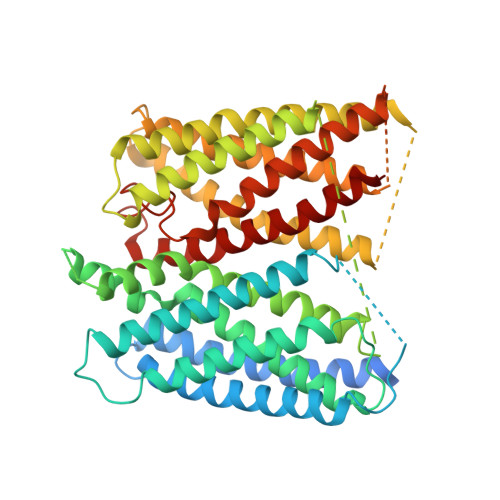
Structures of the otopetrin proton channels Otop1 and Otop3.
Saotome, K., Teng, B., Tsui, C.C.A., Lee, W.H., Tu, Y.H., Kaplan, J.P., Sansom, M.S.P., Liman, E.R., Ward, A.B.(2019) Nat Struct Mol Biol 26: 518-525
- PubMed: 31160780
- DOI: https://doi.org/10.1038/s41594-019-0235-9
- Primary Citation of Related Structures:
6NF4, 6NF6 - PubMed Abstract:
Otopetrins (Otop1-Otop3) comprise one of two known eukaryotic proton-selective channel families. Otop1 is required for otoconia formation and a candidate mammalian sour taste receptor. Here we report cryo-EM structures of zebrafish Otop1 and chicken Otop3 in lipid nanodiscs. The structures reveal a dimeric architecture, with each subunit forming 12 transmembrane helices divided into structurally similar amino (N) and carboxy (C) domains. Cholesterol-like molecules occupy various sites in Otop1 and Otop3 and occlude a central tunnel. In molecular dynamics simulations, hydrophilic vestibules formed by the N and C domains and in the intrasubunit interface between N and C domains form conduits for water entry into the membrane core, suggesting three potential proton conduction pathways. By mutagenesis, we tested the roles of charged residues in each putative permeation pathway. Our results provide a structural basis for understanding selective proton permeation and gating of this conserved family of proton channels.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA.