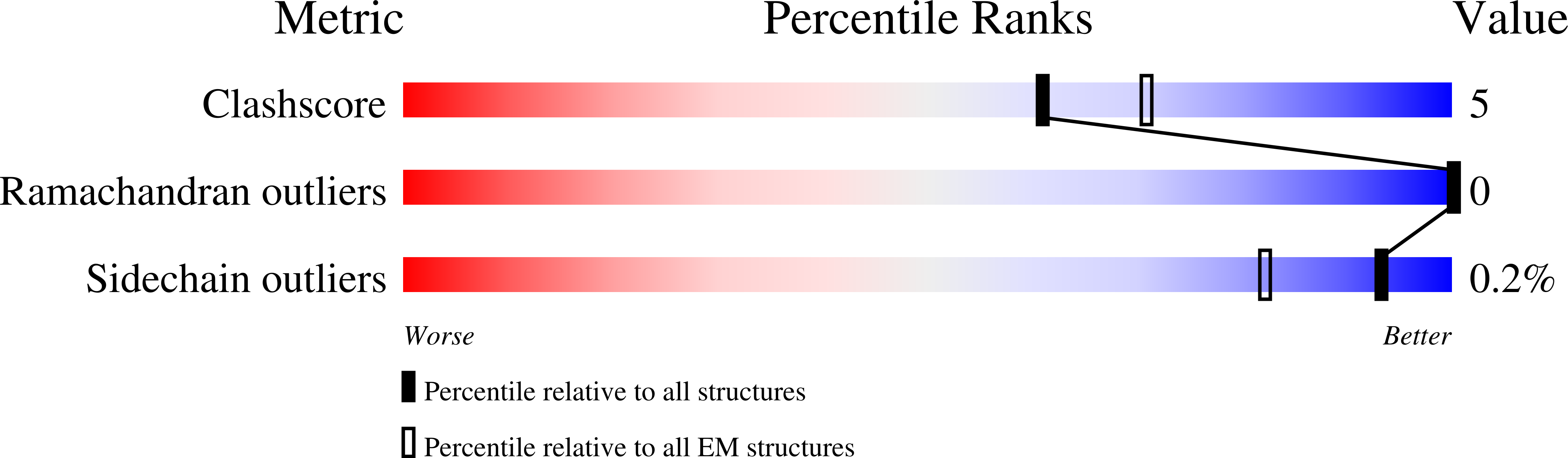Atomic structure of the translation regulatory protein NS1 of bluetongue virus.
Kerviel, A., Ge, P., Lai, M., Jih, J., Boyce, M., Zhang, X., Zhou, Z.H., Roy, P.(2019) Nat Microbiol 4: 837-845
- PubMed: 30778144
- DOI: https://doi.org/10.1038/s41564-019-0369-x
- Primary Citation of Related Structures:
6N9Y - PubMed Abstract:
Bluetongue virus (BTV) non-structural protein 1 (NS1) regulates viral protein synthesis and exists as tubular and non-tubular forms in infected cells, but how tubules assemble and how protein synthesis is regulated are unknown. Here, we report near-atomic resolution structures of two NS1 tubular forms determined by cryo-electron microscopy. The two tubular forms are different helical assemblies of the same NS1 monomer, consisting of an amino-terminal foot, a head and body domains connected to an extended carboxy-terminal arm, which wraps atop the head domain of another NS1 subunit through hydrophobic interactions. Deletion of the C terminus prevents tubule formation but not viral replication, suggesting an active non-tubular form. Two zinc-finger-like motifs are present in each NS1 monomer, and tubules are disrupted by divalent cation chelation and restored by cation addition, including Zn 2+ , suggesting a regulatory role of divalent cations in tubule formation. In vitro luciferase assays show that the NS1 non-tubular form upregulates BTV mRNA translation, whereas zinc-finger disruption decreases viral mRNA translation, tubule formation and virus replication, confirming a functional role for the zinc-fingers. Thus, the non-tubular form of NS1 is sufficient for viral protein synthesis and infectious virus replication, and the regulatory mechanism involved operates through divalent cation-dependent conversion between the non-tubular and tubular forms.
Organizational Affiliation:
Department of Pathogen Molecular Biology, London School of Hygiene and Tropical Medicine, London, UK.