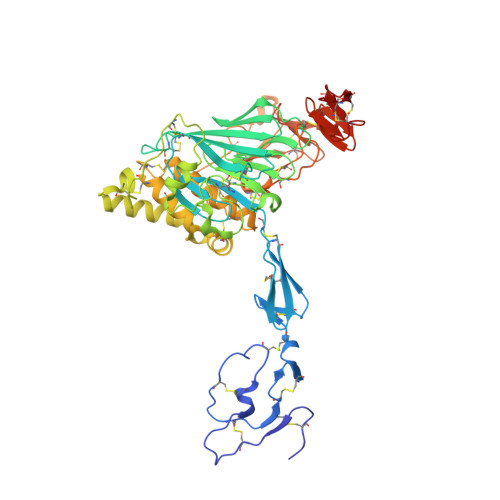
The von Willebrand factor D'D3 assembly and structural principles for factor VIII binding and concatemer biogenesis.
Dong, X., Leksa, N.C., Chhabra, E.S., Arndt, J.W., Lu, Q., Knockenhauer, K.E., Peters, R.T., Springer, T.A.(2019) Blood 133: 1523-1533
- PubMed: 30642920
- DOI: https://doi.org/10.1182/blood-2018-10-876300
- Primary Citation of Related Structures:
6N29 - PubMed Abstract:
D assemblies make up half of the von Willebrand factor (VWF), yet are of unknown structure. D1 and D2 in the prodomain and D'D3 in mature VWF at Golgi pH form helical VWF tubules in Weibel Palade bodies and template dimerization of D3 through disulfides to form ultralong VWF concatemers. D'D3 forms the binding site for factor VIII. The crystal structure of monomeric D'D3 with cysteine residues required for dimerization mutated to alanine was determined at an endoplasmic reticulum (ER)-like pH. The smaller C8-3, TIL3 (trypsin inhibitor-like 3), and E3 modules pack through specific interfaces as they wind around the larger, N-terminal, Ca 2+ -binding von Willebrand D domain (VWD) 3 module to form a wedge shape. D' with its TIL' and E' modules projects away from D3. The 2 mutated cysteines implicated in D3 dimerization are buried, providing a mechanism for protecting them against premature disulfide linkage in the ER, where intrachain disulfide linkages are formed. D3 dimerization requires co-association with D1 and D2, Ca 2+ , and Golgi-like acidic pH. Associated structural rearrangements in the C8-3 and TIL3 modules are required to expose cysteine residues for disulfide linkage. Our structure provides insight into many von Willebrand disease mutations, including those that diminish factor VIII binding, which suggest that factor VIII binds not only to the N-terminal TIL' domain of D' distal from D3 but also extends across 1 side of D3. The organizing principle for the D3 assembly has implications for other D assemblies and the construction of higher-order, disulfide-linked assemblies in the Golgi in both VWF and mucins.
Organizational Affiliation:
Children's Hospital Boston, Boston, MA.