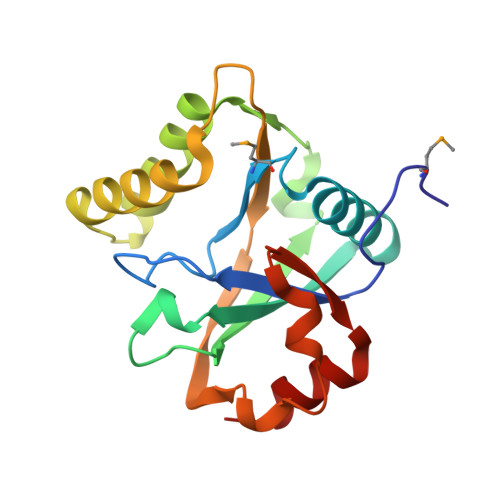
The N-terminal domain of Staphylothermus marinus McrB shares structural homology with PUA-like RNA binding proteins.
Hosford, C.J., Adams, M.C., Niu, Y., Chappie, J.S.(2020) J Struct Biol 211: 107572-107572
- PubMed: 32652237
- DOI: https://doi.org/10.1016/j.jsb.2020.107572
- Primary Citation of Related Structures:
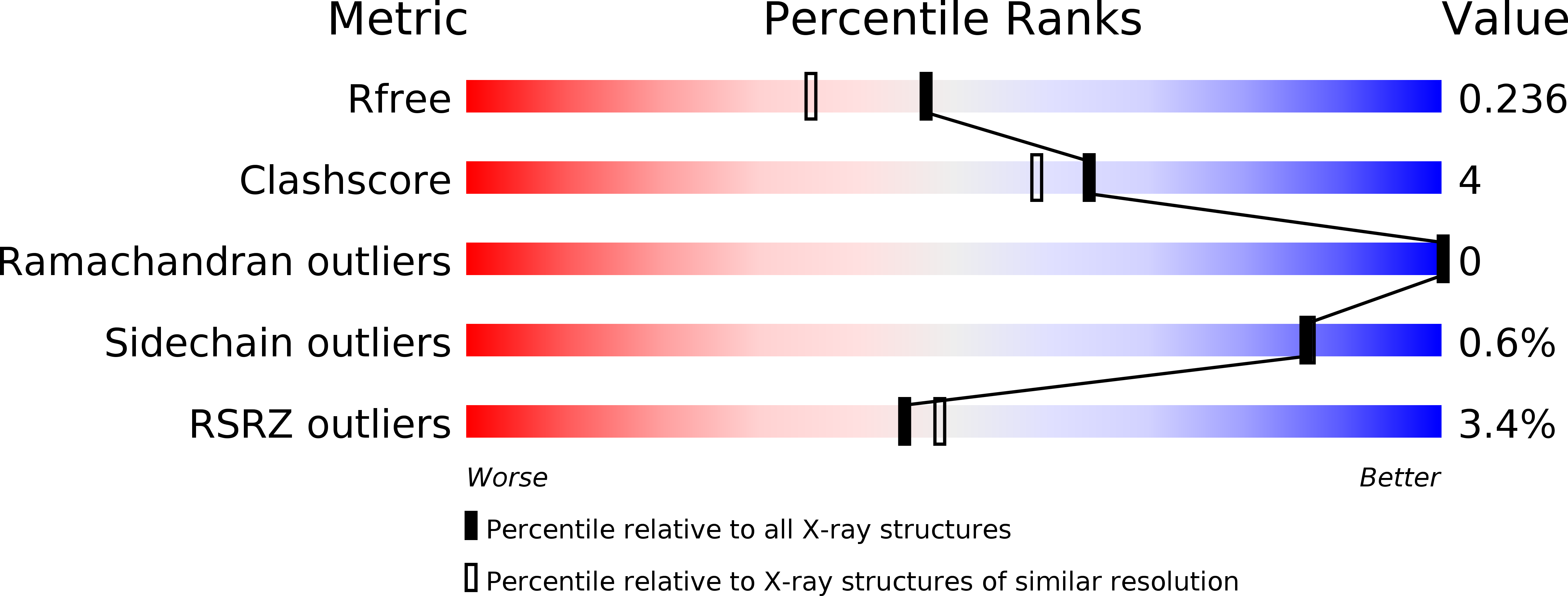
6N0S - PubMed Abstract:
McrBC is a conserved modification-dependent restriction system that in Escherichia coli specifically targets foreign DNA containing methylated cytosines. Crystallographic data show that the N-terminal domain of Escherichia coli McrB binds substrates via a base flipping mechanism. This region is poorly conserved among the plethora of McrB homologs, suggesting that other species may use alternative binding strategies and/or recognize different targets. Here we present the crystal structure of the N-terminal domain from Stayphlothermus marinus McrB (Sm3-180) at 1.92 Å, which adopts a PUA-like EVE fold that is closely related to the YTH and ASCH RNA binding domains. Unlike most PUA-like domains, Sm3-180 binds DNA and can associate with different modified substrates. We find the canonical 'aromatic cage' binding pocket that confers specificity for methylated bases in other EVE/YTH domains is degenerate and occluded in Sm3-180, which may contribute to its promiscuity in target recognition. Further structural comparison between different PUA-like domains identifies motifs and conformational variations that correlate with the preference for binding either DNA or RNA. Together these data have important implications for PUA-like domain specificity and suggest a broader biological versatility for the McrBC family than previously described.
Organizational Affiliation:
Department of Molecular Medicine, Cornell University, Ithaca, NY 14853, USA.