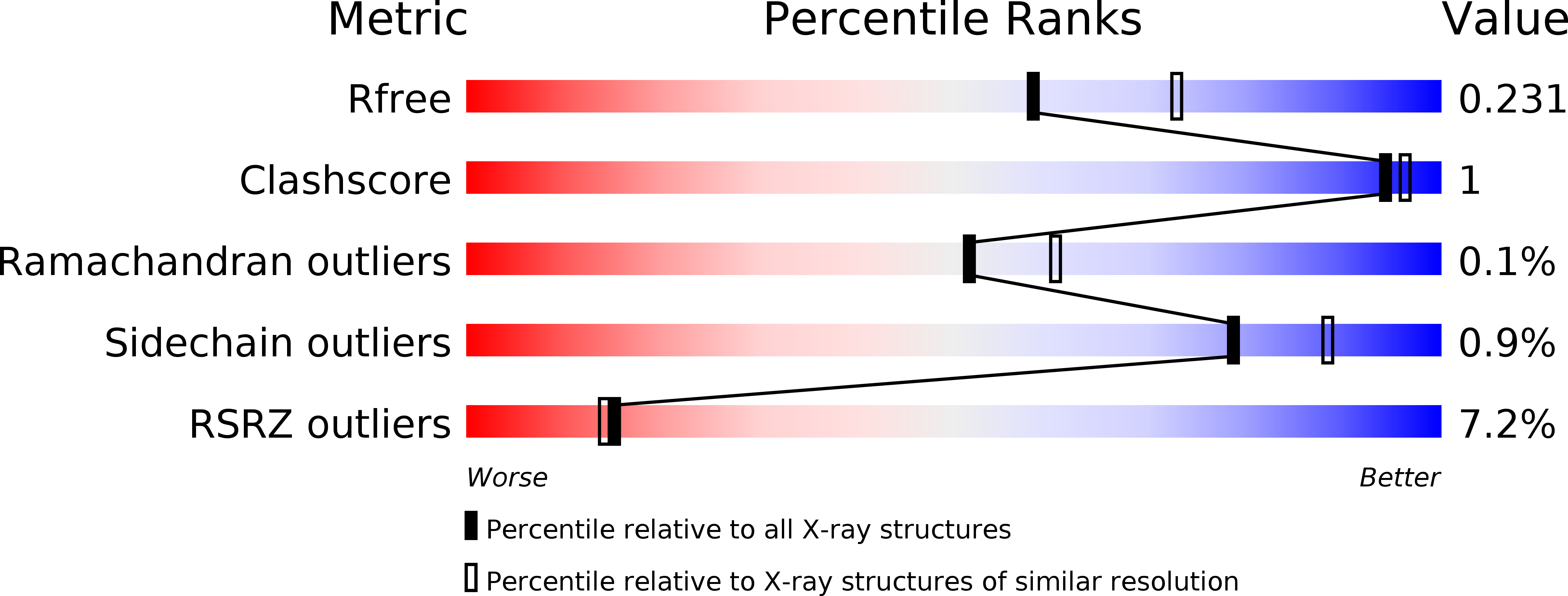
A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1.
Pedersen, L.C., Inoue, K., Kim, S., Perera, L., Shaw, N.D.(2019) Commun Biol 2: 255-255
- PubMed: 31312724
- DOI: https://doi.org/10.1038/s42003-019-0499-y
- Primary Citation of Related Structures:
6MW7 - PubMed Abstract:
Variants in the gene SMCHD1 , which encodes an epigenetic repressor, have been linked to both congenital arhinia and a late-onset form of muscular dystrophy called facioscapulohumeral muscular dystrophy type 2 (FSHD2). This suggests that SMCHD1 has a diversity of functions in both developmental time and space. The C-terminal end of SMCHD1 contains an SMC-hinge domain which mediates homodimerization and chromatin association, whereas the molecular architecture of the N-terminal region, which harbors the GHKL-ATPase domain, is not well understood. We present the crystal structure of the human SMCHD1 N-terminal ATPase module bound to ATP as a functional dimer. The dimer is stabilized by a novel N-terminal ubiquitin-like fold and by a downstream transducer domain. While disease variants map to what appear to be critical interdomain/intermolecular interfaces, only the FSHD2-specific mutant constructs we tested consistently abolish ATPase activity and/or dimerization. These data suggest that the full functional profile of SMCHD1 has yet to be determined.
Organizational Affiliation:
1Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709 USA.