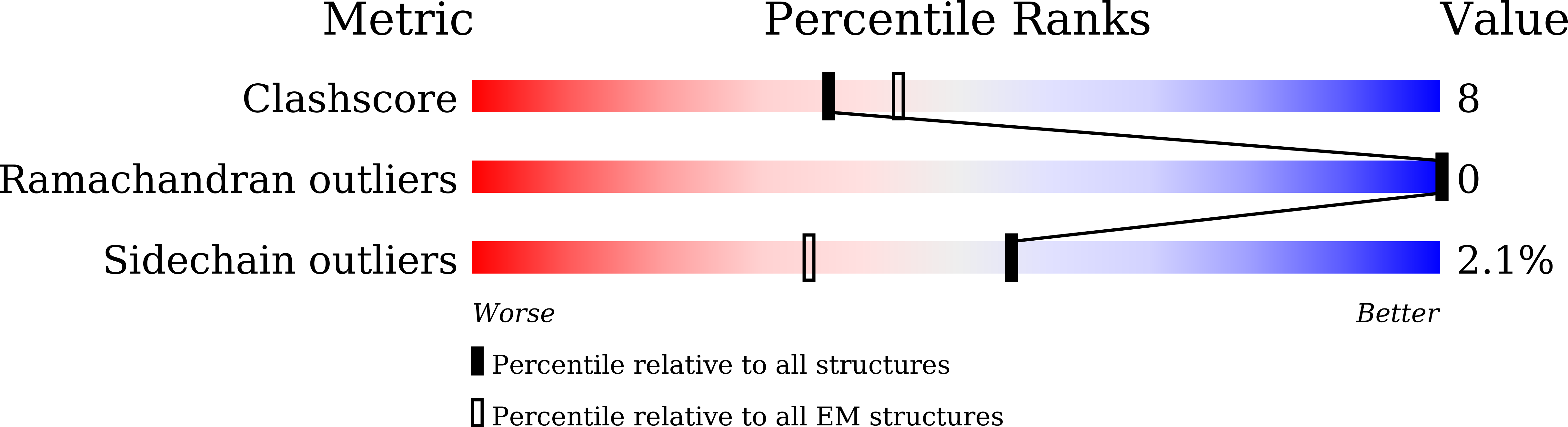Structural Insights into a Plant Mechanosensitive Ion Channel MSL1.
Li, Y., Hu, Y., Wang, J., Liu, X., Zhang, W., Sun, L.(2020) Cell Rep 30: 4518-4527.e3
- PubMed: 32234484
- DOI: https://doi.org/10.1016/j.celrep.2020.03.026
- Primary Citation of Related Structures:
6LYP - PubMed Abstract:
The small conductance mechanosensitive ion channel (MscS)-like (MSL) proteins in plants are evolutionarily conserved homologs of the bacterial small conductance mechanosensitive ion channels. As the sole member of the Arabidopsis MSL family localized in the mitochondrial inner membrane, MSL1 is essential to maintain the normal membrane potential of mitochondria. Here, we report a cryoelectron microscopy (cryo-EM) structure of Arabidopsis thaliana MSL1 (AtMSL1) at 3.3 Å. The overall architecture of AtMSL1 is similar to MscS. However, the transmembrane domain of AtMSL1 is larger. Structural differences are observed in both the transmembrane and the matrix domain of AtMSL1. The carboxyl-terminus of AtMSL1 is more flexible and the β-barrel structure observed in MscS is absent. The side portals in AtMSL1 are significantly smaller, and enlarging the size of the portal by mutagenesis can increase the channel conductance. Our study provides a framework for eukaryotic MscS-like mechanosensitive ion channels and the gating mechanism of the MscS family.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, University of Science and Technology of China, Hefei 230027, China.