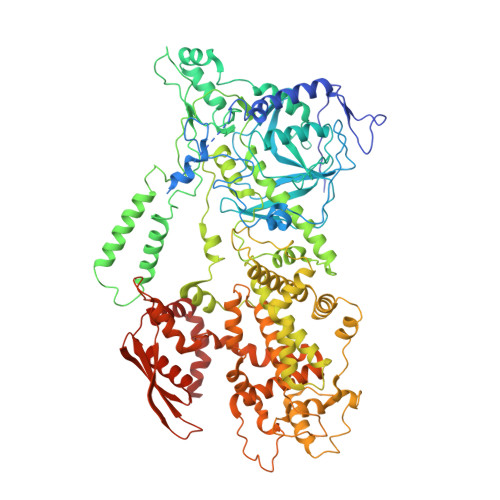
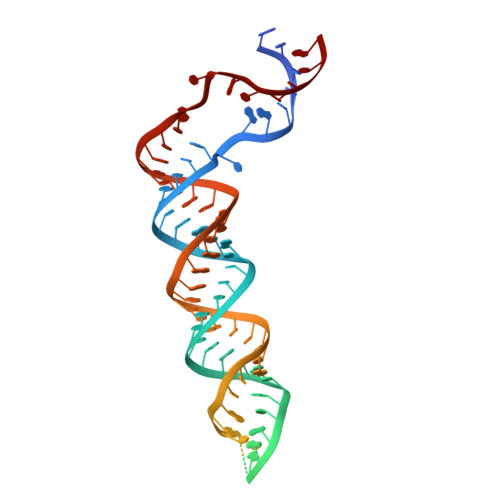
Structural Basis for pri-miRNA Recognition by Drosha.
Jin, W., Wang, J., Liu, C.P., Wang, H.W., Xu, R.M.(2020) Mol Cell 78: 423
- PubMed: 32220645
- DOI: https://doi.org/10.1016/j.molcel.2020.02.024
- Primary Citation of Related Structures:
6LXD, 6LXE - PubMed Abstract:
A commencing and critical step in miRNA biogenesis involves processing of pri-miRNAs in the nucleus by Microprocessor. An important, but not completely understood, question is how Drosha, the catalytic subunit of Microprocessor, binds pri-miRNAs and correctly specifies cleavage sites. Here we report the cryoelectron microscopy structures of the Drosha-DGCR8 complex with and without a pri-miRNA. The RNA-bound structure provides direct visualization of the tertiary structure of pri-miRNA and shows that a helix hairpin in the extended PAZ domain and the mobile basic (MB) helix in the RNase IIIa domain of Drosha coordinate to recognize the single-stranded to double-stranded junction of RNA, whereas the dsRNA binding domain makes extensive contacts with the RNA stem. Furthermore, the RNA-free structure reveals an autoinhibitory conformation of the PAZ helix hairpin. These findings provide mechanistic insights into pri-miRNA cleavage site selection and conformational dynamics governing pri-miRNA recognition by the catalytic component of Microprocessor.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.