Crystal structure of Leptospira leucine-rich repeat 20 reveals a novel E-cadherin binding protein to induce NGAL expression in HK2 cells.
Hsu, S.H., Chu, C.H., Tian, Y.C., Chang, M.Y., Chou, L.F., Sun, Y.J., Yang, C.W.(2020) Biochem J 477: 4313-4326
- PubMed: 33094809
- DOI: https://doi.org/10.1042/BCJ20200547
- Primary Citation of Related Structures:
6LX0 - PubMed Abstract:
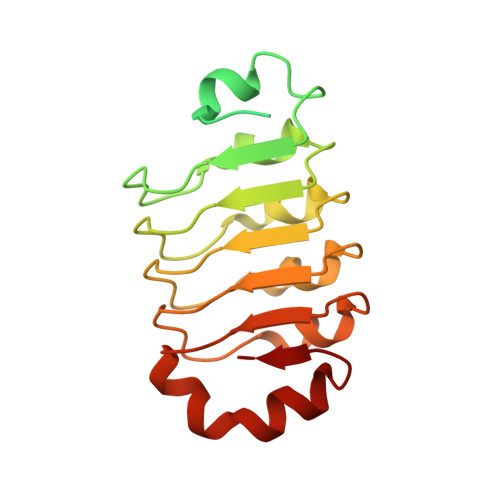
Leptospirosis is the most common zoonotic disease caused by pathogenic Leptospira, which is classified into three groups according to virulence. Its pathogenic and intermediate species contain leucine-rich repeat (LRR) proteins that are rarely expressed in non-pathogenic strains. In this study, we presented the crystal structure of LSS_11580 (rLRR20) from pathogenic L. santarosai serovar Shermani. X-ray diffraction at a resolution of 1.99 Å revealed a horseshoe-shaped structure containing seven α-helices and five β-sheets. Affinity assays indicated that rLRR20 interacts with E-cadherin on the cell surface. Interestingly, its binds to the extracellular (EC) 1 domain in human epithelial (E)-cadherin, which is responsible for binding to another E-cadherin molecule in neighboring cells. Several charged residues on the concave face of LRR20 were predicted to interact with EC1 domain. In the affinity assays, these charged residues were replaced by alanine, and their affinities to E-cadherin were measured. Three vital residues and mutation variants of LRR20, namely D56A, E59A, and E123A, demonstrated significantly reduced affinity to E-cadherin compared with the control. Besides, we also demonstrated that rLRR20 induced the expression of neutrophil gelatinase-associated lipocalin (NGAL) in HK2 cells. The low ability of the three mutation variants to induce NGAL expression further demonstrates this induction. The present findings indicate that LRR20 from pathogenic Leptospira binds to E-cadherin and interacts with its EC1 domain. In addition, its induction of NGAL expression in HK2 cells is associated with acute kidney injury in human.
Organizational Affiliation:
Department of Nephrology, Kidney Research Center, Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Taoyuan 33333, Taiwan.