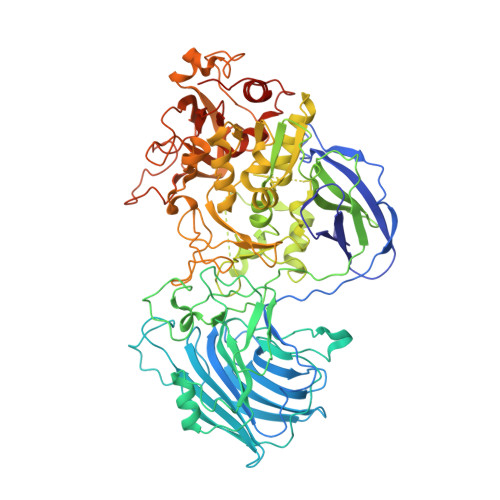
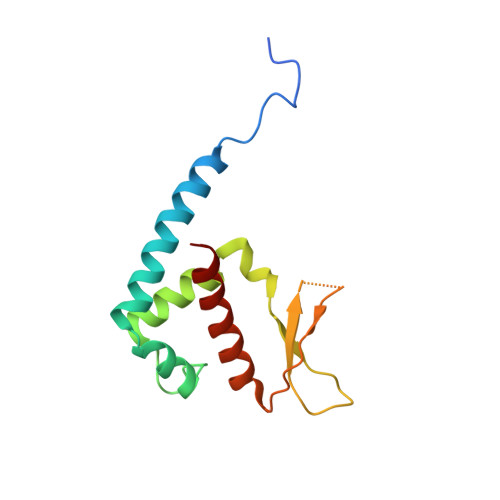
A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Arya, C.K., Yadav, S., Fine, J., Casanal, A., Chopra, G., Ramanathan, G., Vinothkumar, K.R., Subramanian, R.(2020) Angew Chem Int Ed Engl 59: 16961-16966
- PubMed: 32452120
- DOI: https://doi.org/10.1002/anie.202005332
- Primary Citation of Related Structures:
6LVB, 6LVC, 6LVD, 6LVE, 6LVV - PubMed Abstract:
N,N-dimethyl formamide (DMF) is an extensively used organic solvent but is also a potent pollutant. Certain bacterial species from genera such as Paracoccus, Pseudomonas, and Alcaligenes have evolved to use DMF as a sole carbon and nitrogen source for growth via degradation by a dimethylformamidase (DMFase). We show that DMFase from Paracoccus sp. strain DMF is a halophilic and thermostable enzyme comprising a multimeric complex of the α 2 β 2 or (α 2 β 2 ) 2 type. One of the three domains of the large subunit and the small subunit are hitherto undescribed protein folds of unknown evolutionary origin. The active site consists of a mononuclear iron coordinated by two Tyr side-chain phenolates and one carboxylate from Glu. The Fe 3+ ion in the active site catalyzes the hydrolytic cleavage of the amide bond in DMF. Kinetic characterization reveals that the enzyme shows cooperativity between subunits, and mutagenesis and structural data provide clues to the catalytic mechanism.
Organizational Affiliation:
Department of Chemistry, Indian Institute of Technology, Kanpur, India.