Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
He, H.P., Luo, M., Cao, Y.L., Lin, Y.X., Zhang, H., Zhang, X., Ou, J.Y., Yu, B., Chen, X., Xu, M., Feng, L., Zeng, M.S., Zeng, Y.X., Gao, S.(2020) Nat Commun 11: 5405-5405
- PubMed: 33106493
- DOI: https://doi.org/10.1038/s41467-020-19259-x
- Primary Citation of Related Structures:
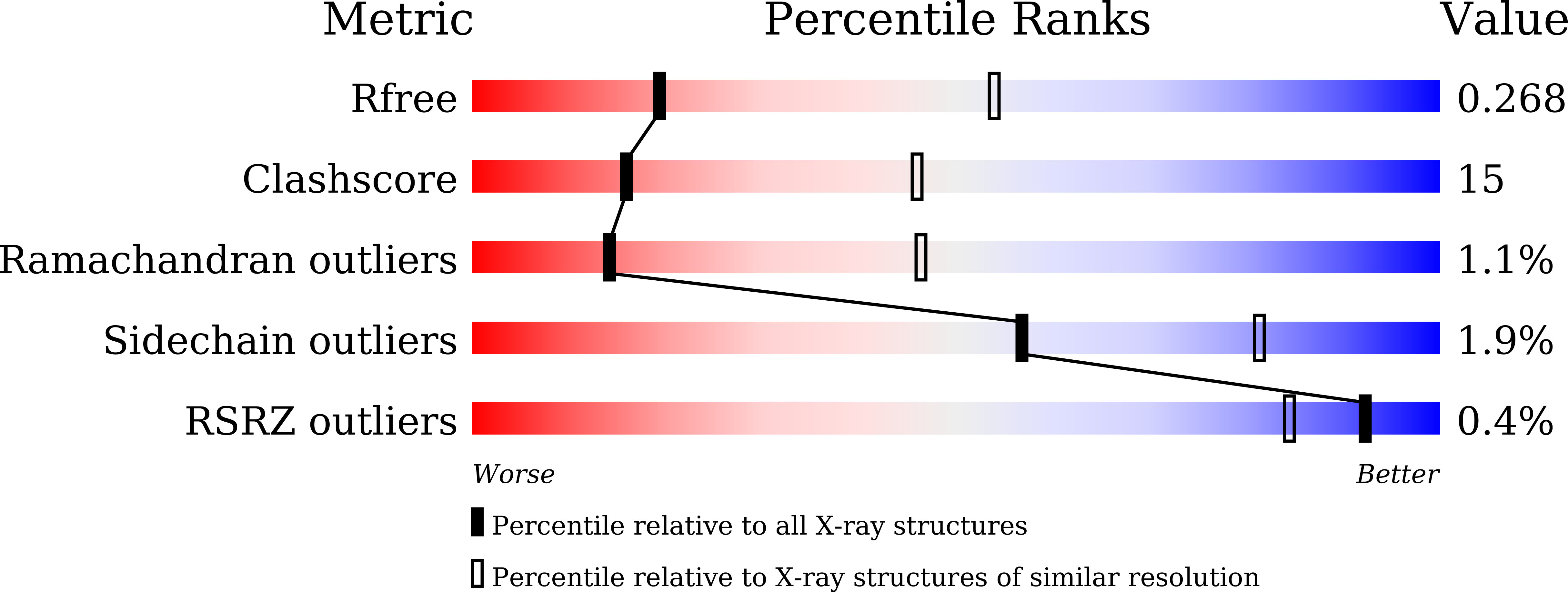
6LQN, 6LQO - PubMed Abstract:
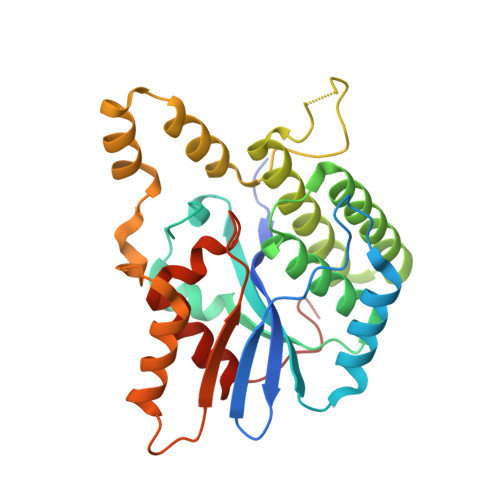
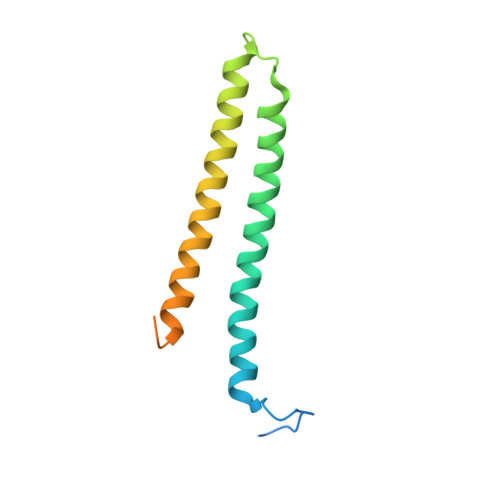
Epstein-Barr virus (EBV) is a γ-herpesvirus associated with the occurrence of several human malignancies. BBRF2 and BSRF1 are two EBV tegument proteins that have been suggested to form a hetero-complex and mediate viral envelopment, but the molecular basis of their interaction and the functional mechanism of this complex remains unknown. Here, we present crystal structures of BBRF2 alone and in complex with BSRF1. BBRF2 has a compact globular architecture featuring a central β-sheet that is surrounded by 10 helices, it represents a novel fold distinct from other known protein structures. The central portion of BSRF1 folds into two tightly associated antiparallel α-helices, forming a composite four-helix bundle with two α-helices from BBRF2 via a massive hydrophobic network. In vitro, a BSRF1-derived peptide binds to BBRF2 and reduces the number of viral genome copies in EBV-positive cells. Exogenous BBRF2 and BSRF1 co-localize at the Golgi apparatus. Furthermore, BBRF2 binds capsid and capsid-associated proteins, whereas BSRF1 associates with glycoproteins. These findings indicate that the BBRF2-BSRF1 complex tethers EBV nucleocapsids to the glycoprotein-enriched Golgi membrane, facilitating secondary envelopment.
Organizational Affiliation:
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, 510060, Guangzhou, China.