Biochemical Characterization and Crystal Structure of a Novel NAD+ -Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum.
Huang, S.P., Zhou, L.C., Wen, B., Wang, P., Zhu, G.P.(2020) Int J Mol Sci 21
- PubMed: 32824636
- DOI: https://doi.org/10.3390/ijms21165915
- Primary Citation of Related Structures:
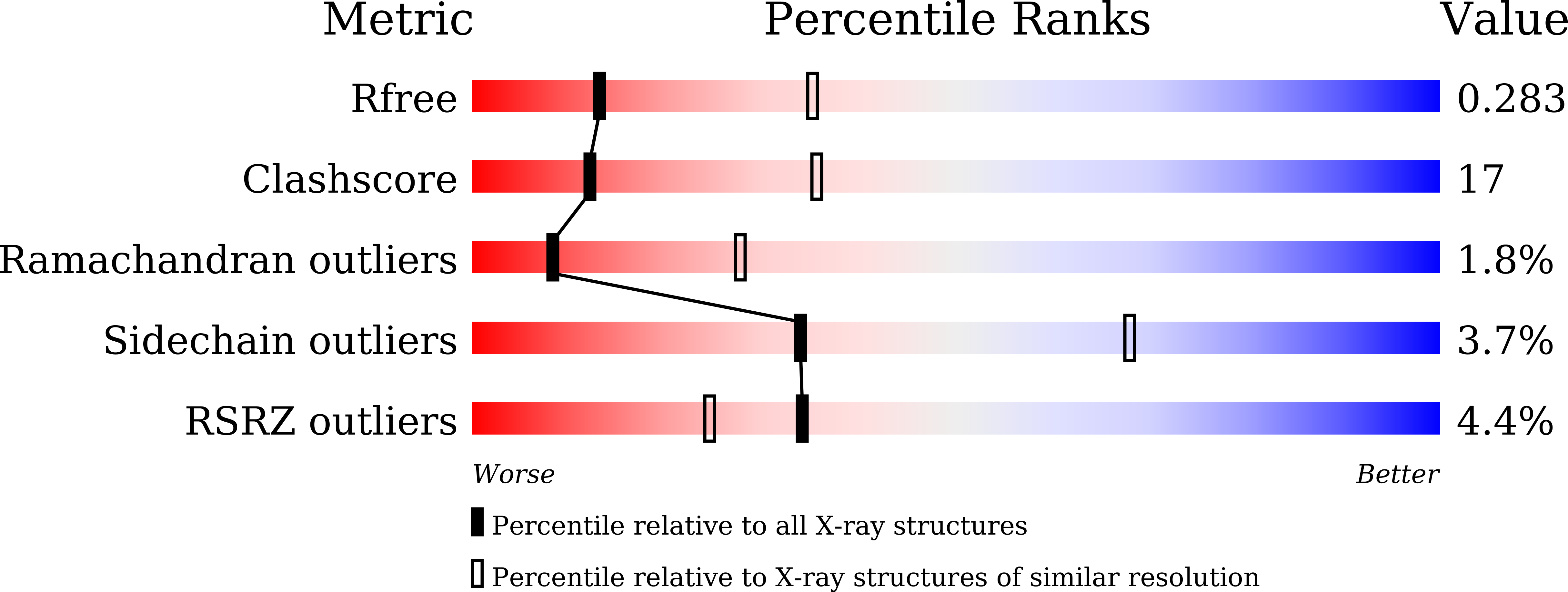
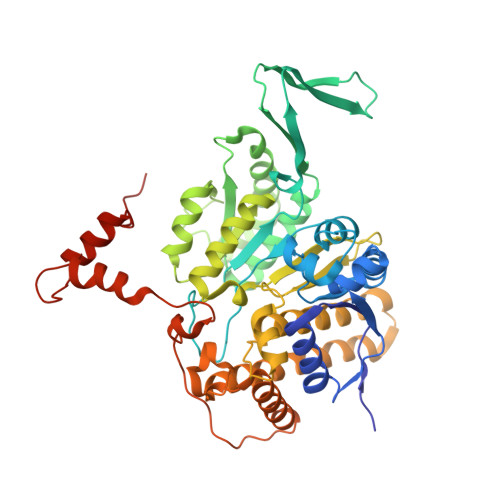
6LKZ - PubMed Abstract:
The marine diatom Phaeodactylum tricornutum originated from a series of secondary symbiotic events and has been used as a model organism for studying diatom biology. A novel type II homodimeric isocitrate dehydrogenase from P. tricornutum (PtIDH1) was expressed, purified, and identified in detail through enzymatic characterization. Kinetic analysis showed that PtIDH1 is NAD + -dependent and has no detectable activity with NADP + . The catalytic efficiency of PtIDH1 for NAD + is 0.16 μM -1 ·s -1 and 0.09 μM -1 ·s -1 in the presence of Mn 2+ and Mg 2+ , respectively. Unlike other bacterial homodimeric NAD-IDHs, PtIDH1 activity was allosterically regulated by the isocitrate. Furthermore, the dimeric structure of PtIDH1 was determined at 2.8 Å resolution, and each subunit was resolved into four domains, similar to the eukaryotic homodimeric NADP-IDH in the type II subfamily. Interestingly, a unique and novel C-terminal EF-hand domain was first defined in PtIDH1. Deletion of this domain disrupted the intact dimeric structure and activity. Mutation of the four Ca 2+ -binding sites in the EF-hand significantly reduced the calcium tolerance of PtIDH1. Thus, we suggest that the EF-hand domain could be involved in the dimerization and Ca 2+ -coordination of PtIDH1. The current report, on the first structure of type II eukaryotic NAD-IDH, provides new information for further investigation of the evolution of the IDH family.
Organizational Affiliation:
Anhui Provincial Key Laboratory of Molecular Enzymology and Mechanism of Major Diseases and Key Laboratory of Biomedicine in Gene Diseases and Health of Anhui Higher Education Institutes, College of Life Sciences, Anhui Normal University, Wuhu 241000, China.