Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Zhang, Q., Cao, H.Y., Wei, L., Lu, D., Du, M., Yuan, M., Shi, D., Chen, X., Wang, P., Chen, X.L., Chi, L., Zhang, Y.Z., Li, F.(2021) Nat Commun 12: 1263-1263
- PubMed: 33627653
- DOI: https://doi.org/10.1038/s41467-021-21441-8
- Primary Citation of Related Structures:
6LJA, 6LJL - PubMed Abstract:
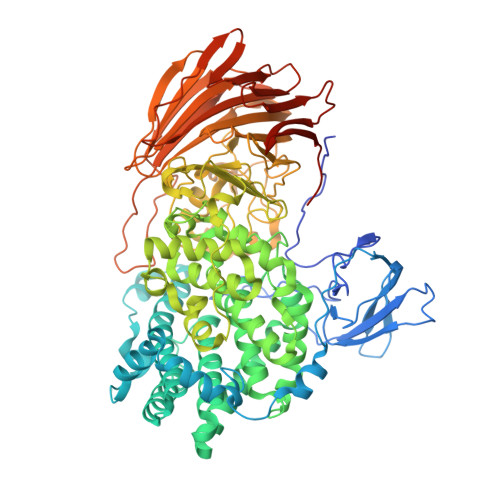
Heparinases (Hepases) are critical tools for the studies of highly heterogeneous heparin (HP)/heparan sulfate (HS). However, exolytic heparinases urgently needed for the sequencing of HP/HS chains remain undiscovered. Herein, a type of exolytic heparinases (exoHepases) is identified from the genomes of different bacteria. These exoHepases share almost no homology with known Hepases and prefer to digest HP rather than HS chains by sequentially releasing unsaturated disaccharides from their reducing ends. The structural study of an exoHepase (BIexoHep) shows that an N-terminal conserved DUF4962 superfamily domain is essential to the enzyme activities of these exoHepases, which is involved in the formation of a unique L-shaped catalytic cavity controlling the sequential digestion of substrates through electrostatic interactions. Further, several HP octasaccharides have been preliminarily sequenced by using BIexoHep. Overall, this study fills the research gap of exoHepases and provides urgently needed tools for the structural and functional studies of HP/HS chains.
Organizational Affiliation:
National Glycoengineering Research Center and Shandong Key Laboratory of Carbohydrate Chemistry and Glycobiology, Shandong University, Qingdao, China.