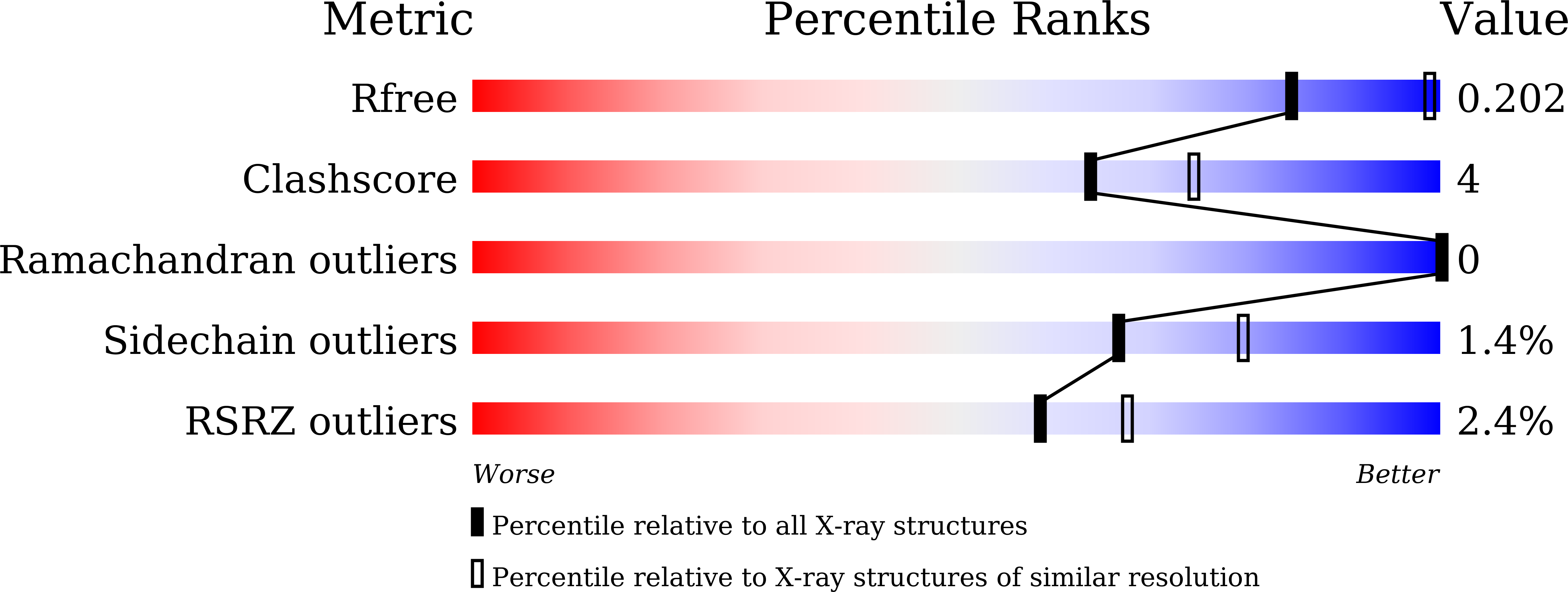
Catalytic specificity of the Lactobacillus plantarum cystathionine gamma-lyase presumed by the crystallographic analysis.
Matoba, Y., Noda, M., Yoshida, T., Oda, K., Ezumi, Y., Yasutake, C., Izuhara-Kihara, H., Danshiitsoodol, N., Kumagai, T., Sugiyama, M.(2020) Sci Rep 10: 14886-14886
- PubMed: 32913258
- DOI: https://doi.org/10.1038/s41598-020-71756-7
- Primary Citation of Related Structures:
6LDO, 6LE4 - PubMed Abstract:
The reverse transsulfuration pathway, which is composed of cystathionine β-synthase (CBS) and cystathionine γ-lyase (CGL), plays a role to synthesize L-cysteine using L-serine and the sulfur atom in L-methionine. A plant-derived lactic acid bacterium Lactobacillus plantarum SN35N has been previously found to harbor the gene cluster encoding the CBS- and CGL-like enzymes. In addition, it has been demonstrated that the L. plantarum CBS can synthesize cystathionine from O-acetyl-L-serine and L-homocysteine. The aim of this study is to characterize the enzymatic functions of the L. plantarum CGL. We have found that the enzyme has the high γ-lyase activity toward cystathionine to generate L-cysteine, together with the β-lyase activity toward L-cystine to generate L-cysteine persulfide. By the crystallographic analysis of the inactive CGL K194A mutant complexed with cystathionine, we have found the residues which recognize the distal amino and carboxyl groups of cystathionine or L-cystine. The PLP-bound substrates at the active site may take either the binding pose for the γ- or β-elimination reaction, with the former being the major reaction in the case of cystathionine.
Organizational Affiliation:
Faculty of Pharmacy, Yasuda Women's University, Yasuhigashi 6-13-1, Asaminami-ku, Hiroshima, 731-0153, Japan. matoba@yasuda-u.ac.jp.