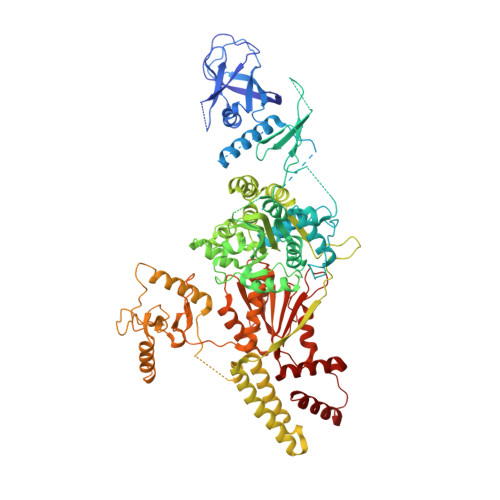
Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Shen, M., Dhingra, N., Wang, Q., Cheng, C., Zhu, S., Tian, X., Yu, J., Gong, X., Li, X., Zhang, H., Xu, X., Zhai, L., Xie, M., Gao, Y., Deng, H., He, Y., Niu, H., Zhao, X., Xiang, S.(2021) Nat Commun 12: 321-321
- PubMed: 33436623
- DOI: https://doi.org/10.1038/s41467-020-20538-w
- Primary Citation of Related Structures:
6L8N, 6L8O - PubMed Abstract:
The yeast protein Rad5 and its orthologs in other eukaryotes promote replication stress tolerance and cell survival using their multiple activities, including ubiquitin ligase, replication fork remodeling and DNA lesion targeting activities. Here, we present the crystal structure of a nearly full-length Rad5 protein. The structure shows three distinct, but well-connected, domains required for Rad5's activities. The spatial arrangement of these domains suggest that different domains can have autonomous activities but also undergo intrinsic coordination. Moreover, our structural, biochemical and cellular studies demonstrate that Rad5's HIRAN domain mediates interactions with the DNA metabolism maestro factor PCNA and contributes to its poly-ubiquitination, binds to DNA and contributes to the Rad5-catalyzed replication fork regression, defining a new type of HIRAN domains with multiple activities. Our work provides a framework to understand how Rad5 integrates its various activities in replication stress tolerance.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Tianjin Medical University, 300070, Tianjin, P. R. China.