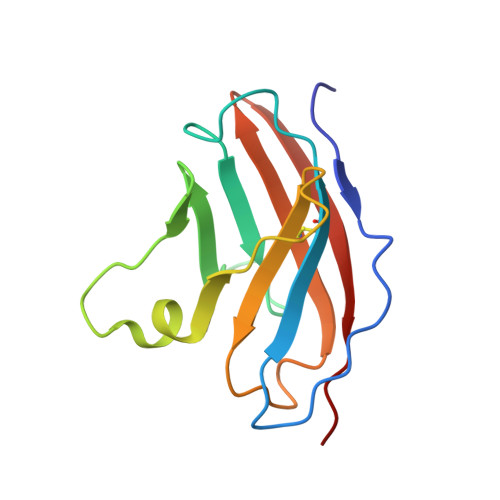
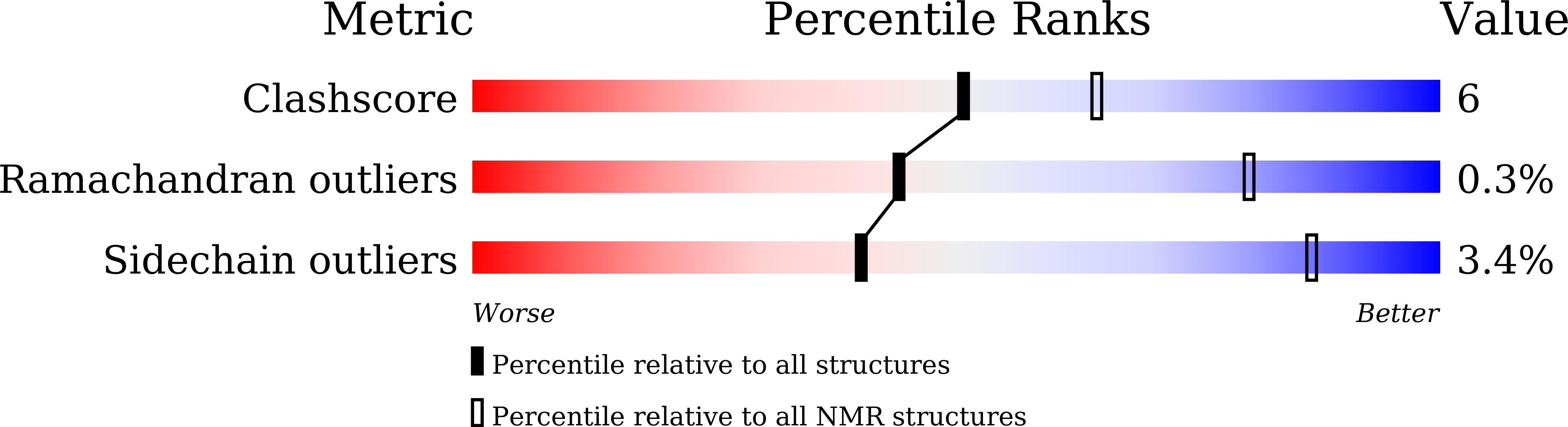Structural Insights into N-terminal IgV Domain of BTNL2, a T Cell Inhibitory Molecule, Suggests a Non-canonical Binding Interface for Its Putative Receptors.
Basak, A.J., Maiti, S., Hansda, A., Mahata, D., Duraivelan, K., Kundapura, S.V., Lee, W., Mukherjee, G., De, S., Samanta, D.(2020) J Mol Biol 432: 5938-5950
- PubMed: 32976909
- DOI: https://doi.org/10.1016/j.jmb.2020.09.013
- Primary Citation of Related Structures:
6L7Z - PubMed Abstract:
T cell costimulation is mediated by the interaction of a number of receptors and ligands present on the surface of the T cell and antigen-presenting cell, respectively. Stimulatory or inhibitory signals from these receptor-ligand interactions work in tandem to preserve immune homeostasis. BTNL2 is a type-1 membrane protein that provides inhibitory signal to T cells and plays an important role in several inflammatory and autoimmune diseases. Therefore, manipulation of the molecular interaction of BTNL2 with its putative receptor could provide strategies to restore immune homeostasis in these diseases. Hence, it is imperative to study the structural characteristics of this molecule, which will provide important insights into its function as well. In this study, the membrane-distal ectodomain of murine BTNL2 was expressed in bacteria as inclusion bodies, refolded in vitro and purified for functional and structural characterization. The domain is monomeric in solution as demonstrated by size-exclusion chromatography and analytical ultracentrifugation, and also binds to its putative receptor on naïve B cells and activated T cell subsets. Importantly, for the first time, we report the structure of BTNL2 as determined by solution NMR spectroscopy and also the picosecond-nanosecond timescale backbone dynamics of this domain. The N-terminal ectodomain of BTNL2, which was able to inhibit T cell function as well, exhibits distinctive structural features. The N-terminal ectodomain of BTNL2 has a significantly reduced surface area in the front sheet due to the non-canonical conformation of the CC' loop, which provides important insights into the recognition of its presently unknown binding partner.
Organizational Affiliation:
School of Bioscience, Indian Institute of Technology Kharagpur, India.