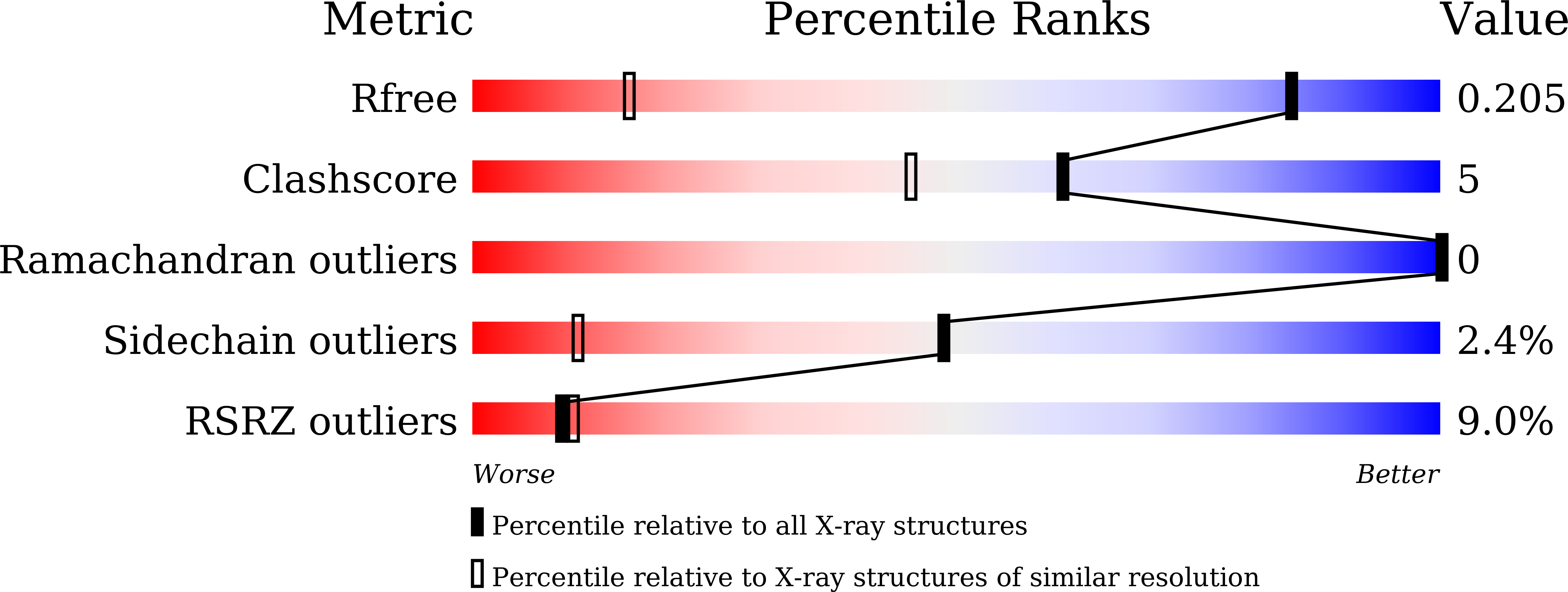
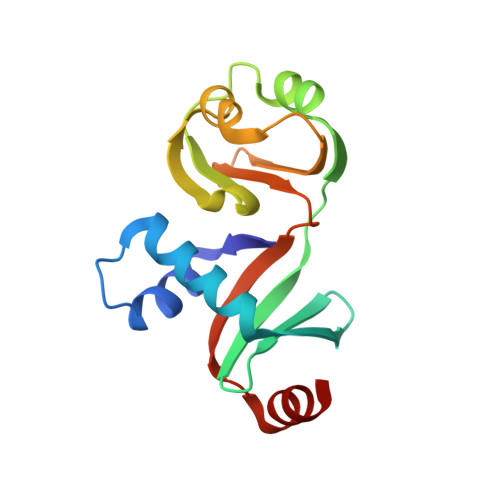
Crystal structure of PYCH_01220 from Pyrococcus yayanosii potentially involved in binding nucleic acid.
Noh, H., Jeon, J.H., Kim, Y.G., Oh, B.H.(2021) Proteins 89: 468-472
- PubMed: 33236809
- DOI: https://doi.org/10.1002/prot.26029
- Primary Citation of Related Structures:
6L7Q - PubMed Abstract:
We report the crystal structure of PYCH_01220, a hypothetical protein in Pyrococcus yayanosii CH1. This protein is composed of two domains, named Domain A and Domain B. While Domain B is not significantly homologous to known protein structures, Domain A is structurally analogous to the C-terminal ribonuclease domain of Escherichia coli colicin D. Domain A has a positively charged surface patch rendered by 13 basic residues, eight arginine or lysine residues of which are evolutionarily conserved. Electrophoretic mobility shift assays showed that PYCH_01220 binds to DNA, and charge-inversion mutations on this patch negatively affect the DNA binding, suggesting that the function of PYCH_01220 might involve nucleic acid-binding via the positively charged patch.
Organizational Affiliation:
Department of Biological Sciences, KAIST Institute for the Biocentury, Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea.