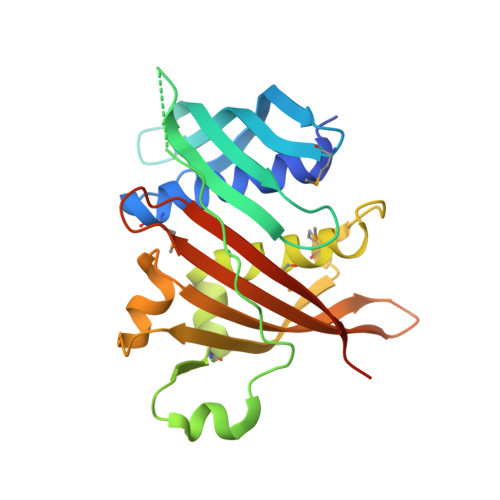
Structural analysis of the meiosis-related protein MS5 reveals non-canonical papain enhancement by cystatin-like folds.
Wang, X., Gao, Y., Guan, Z., Xie, Z., Zhang, D., Yin, P., Yang, G., Hong, D., Xin, Q.(2020) FEBS Lett 594: 2462-2471
- PubMed: 32415887
- DOI: https://doi.org/10.1002/1873-3468.13817
- Primary Citation of Related Structures:
6L77 - PubMed Abstract:
MS5 is a meiosis-related protein belonging to the Brassicaceae-specific domain of unknown function family and characterized by the MS5 superfamily domain (MSD). In this study, we elucidated the three-dimensional crystal structure and potential biochemical function of the MSD. It was observed that the MSD adopts a cystatin-like fold, mainly consisting of a central α-helix and four- or five-stranded antiparallel β-sheets that wrap around it. However, unlike cystatins, which inhibit cysteine proteases, the MSD displayed allosteric activation of papain. We believe that our study provides insight into novel mechanisms of proteolytic enzyme regulation and may serve as a basis for functional studies of the MS5 family proteins in plants.
Organizational Affiliation:
National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China.