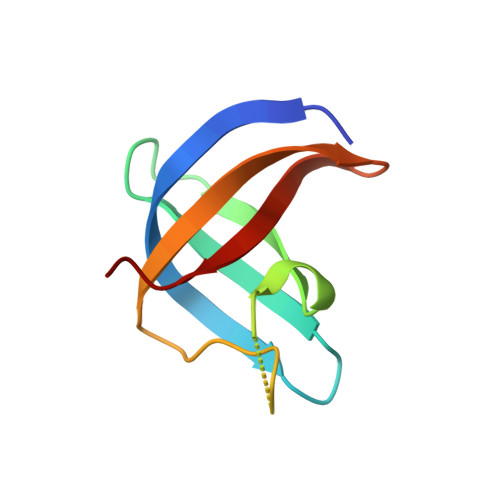
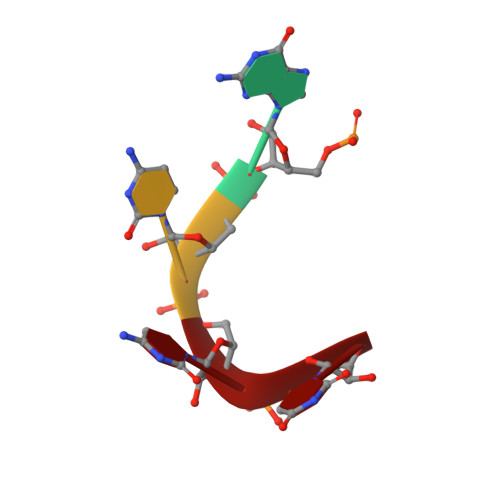
DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Zou, F., Tu, R., Duan, B., Yang, Z., Ping, Z., Song, X., Chen, S., Price, A., Li, H., Scott, A., Perera, A., Li, S., Xie, T.(2020) Proc Natl Acad Sci U S A 117: 3603-3609
- PubMed: 32015133
- DOI: https://doi.org/10.1073/pnas.1910862117
- Primary Citation of Related Structures:
6KTC, 6KUG - PubMed Abstract:
5-Methylcytosine (m 5 C) is a RNA modification that exists in tRNAs and rRNAs and was recently found in mRNAs. Although it has been suggested to regulate diverse biological functions, whether m 5 C RNA modification influences adult stem cell development remains undetermined. In this study, we show that Ypsilon schachtel (YPS), a homolog of human Y box binding protein 1 (YBX1), promotes germ line stem cell (GSC) maintenance, proliferation, and differentiation in the Drosophila ovary by preferentially binding to m 5 C-containing RNAs. YPS is genetically demonstrated to function intrinsically for GSC maintenance, proliferation, and progeny differentiation in the Drosophila ovary, and human YBX1 can functionally replace YPS to support normal GSC development. Highly conserved cold-shock domains (CSDs) of YPS and YBX1 preferentially bind to m 5 C RNA in vitro. Moreover, YPS also preferentially binds to m 5 C-containing RNAs, including mRNAs, in germ cells. The crystal structure of the YBX1 CSD-RNA complex reveals that both hydrophobic stacking and hydrogen bonds are critical for m 5 C binding. Overexpression of RNA-binding-defective YPS and YBX1 proteins disrupts GSC development. Taken together, our findings show that m 5 C RNA modification plays an important role in adult stem cell development.
Organizational Affiliation:
Harbin Institute of Technology, 150001 Heilongjiang, China.