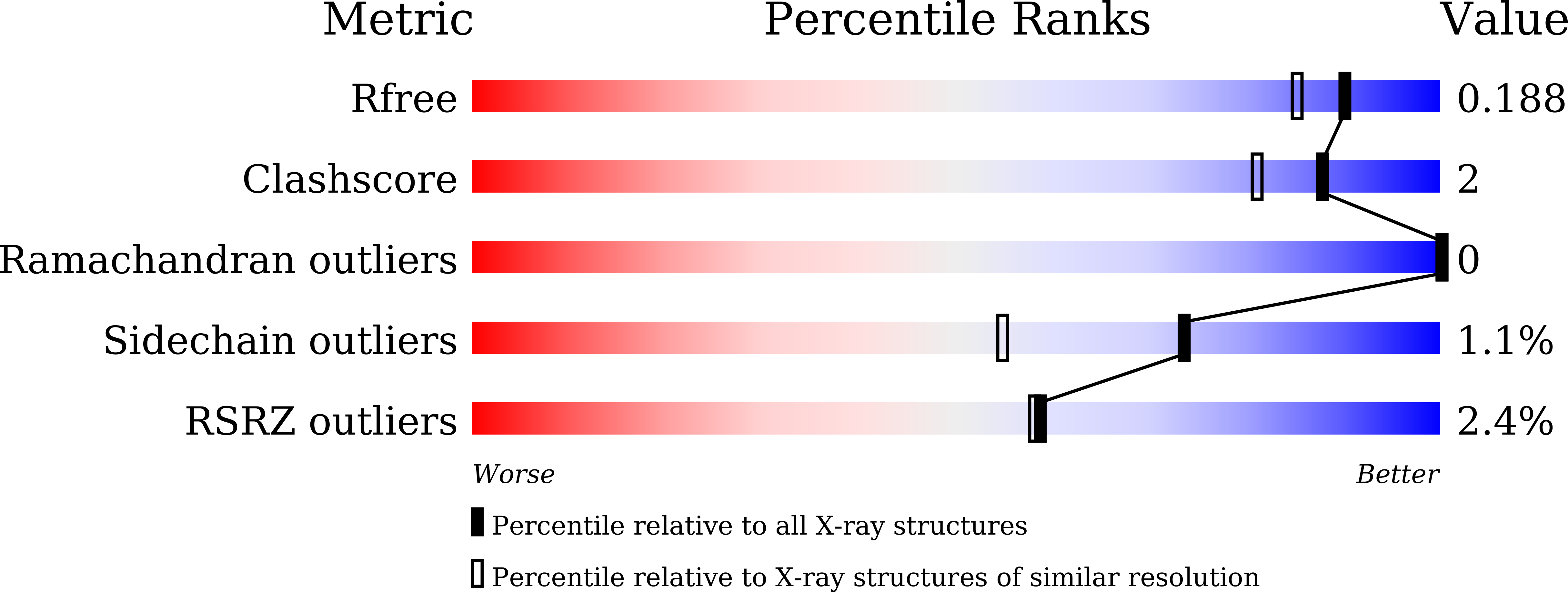
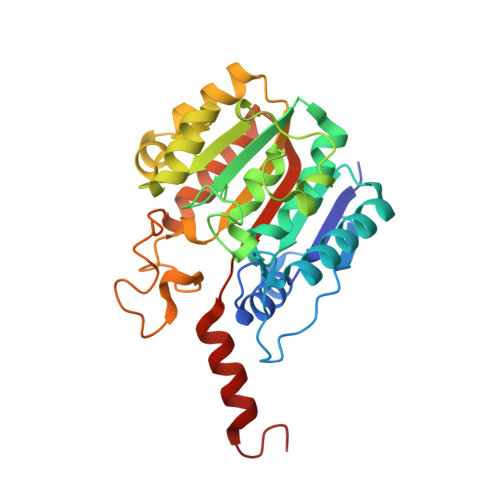
Characterization of a Dimeric Arginase FromZymomonas mobilisZM4.
Hwangbo, S.A., Kim, J.W., Jung, S.J., Jin, K.S., Lee, J.O., Kim, J.S., Park, S.Y.(2019) Front Microbiol 10: 2755-2755
- PubMed: 32038508
- DOI: https://doi.org/10.3389/fmicb.2019.02755
- Primary Citation of Related Structures:
6KSY - PubMed Abstract:
Many organisms have genes to protect themselves from toxic conditions such as high ethanol and/or ammonia concentrations. When a high ethanol condition is induced to Zymomonas mobilis ZM4, a representative ethanologenic organism, this bacterium overexpresses several genes to overcome this ethanol stress. Among them, we characterized a gene product annotated as an arginase (zmARG) from Z. mobilis ZM4. Even though all of the arginase-determining sequence motifs are not strictly conserved in zmARG, this enzyme converts L-arginine to urea and L-ornithine in the presence of a divalent manganese ion. The revealed high-resolution crystal structure of zmARG shows that it has a typical globular α/β arginase fold with a protruded C-terminal helix. Two zinc ions reside in the active site, where one metal ion is penta-coordinated and the other has six ligands, discerning this zmARG from the reported arginases with two hexa-liganded metal ions. zmARG forms a dimeric structure in solution as well as in the crystalline state. The dimeric assembly of zmARG is formed mainly by interaction formed between the C-terminal α-helix of one molecule and the α/β hydrolase fold of another molecule. The presented findings demonstrate the first reported dimeric arginase formed by the C-terminal tail and has two metal ions coordinated by different number of ligands.
Organizational Affiliation:
Pohang Accelerator Laboratory, Pohang University of Science and Technology, Pohang, South Korea.