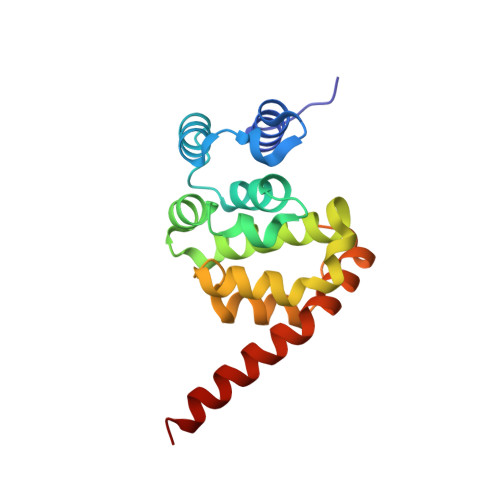
The pH-dependent conformational change of eukaryotic translation initiation factor 5: Insights into partner-binding manner.
Ye, Y., Chen, M., Kato, K., Yao, M.(2019) Biochem Biophys Res Commun 519: 186-191
- PubMed: 31492496
- DOI: https://doi.org/10.1016/j.bbrc.2019.08.128
- Primary Citation of Related Structures:
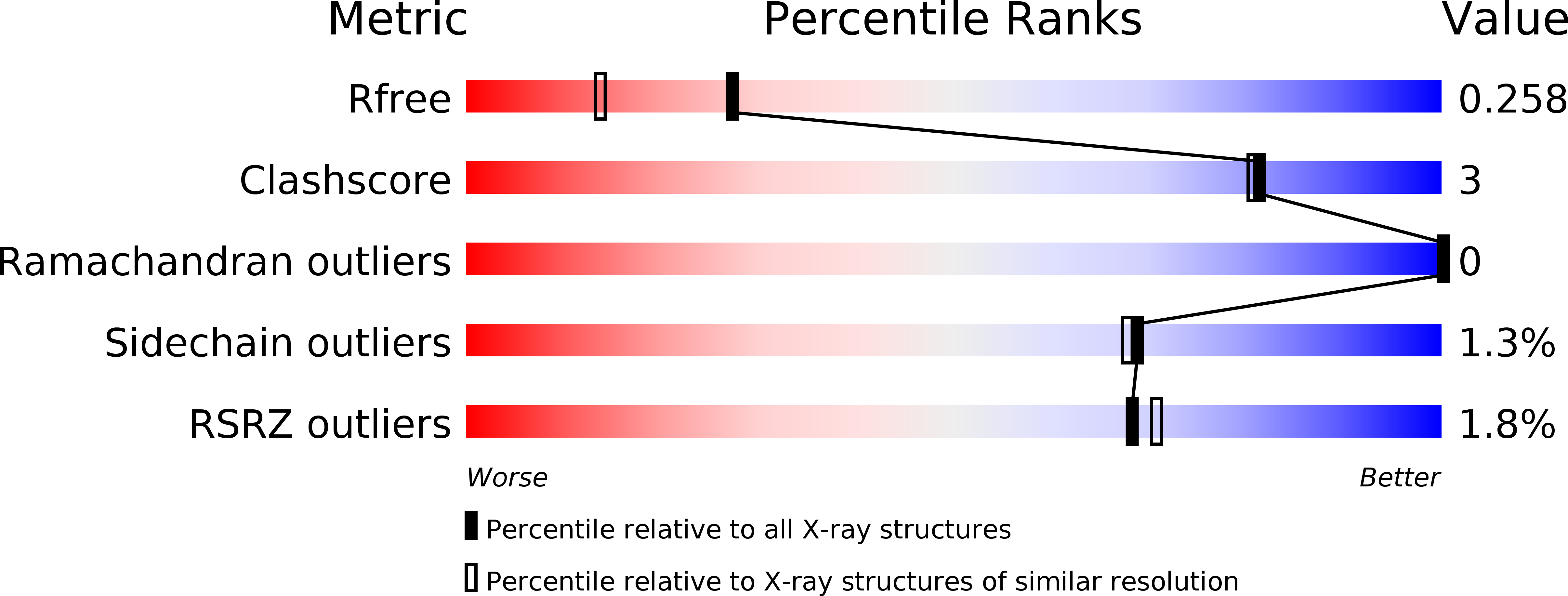
6KND, 6KNE - PubMed Abstract:
In the process of eukaryotic translation, the formation of preinitiation complex 43S, which consists of a 40S subunit, the eIF2-GTP-Met-tRNAi Met ternary complex, eIF3, eIF1, eIF1A, and eIF5, is essential for translational quality control. Of those factors, eIF5 promotes the hydrolysis of eIF2-bound GTP to release eIF2-GDP in the complex for the recycling of eIF2. eIF5 appears to bind to the β subunit of eIF2 (eIF2β) via an interaction between aromatic/acidic residue-rich regions (AA-boxes) in the C-terminal domain of eIF5 (eIF5CTD) and three lysine clusters (K-boxes) in the N-terminal domain of eIF2β (eIF2βNTD). However, the details of this interaction are unclear, due to the lack of a structure for the eIF5-eIF2β complex, and the unavailability of an intact structure of eIF5, in which the AA-boxes are always disordered, with high flexibility. In this study, we solved two crystal structures of eIF5CTD from Candida albicans, which for the first time showed the AA-box2 of eIF5 presenting as an ordered helical structure. The structures exhibited different arrangements of AA-box2 under different pH values, which may reflect the dynamic nature of the interactions of eIF5CTD, and eIF2βNTD in the preinitiation complex.
Organizational Affiliation:
Faculty of Advanced Life Science, Hokkaido University, Kita-10, Nishi-8, Kita-ku, Sapporo, 060-0810, Japan.