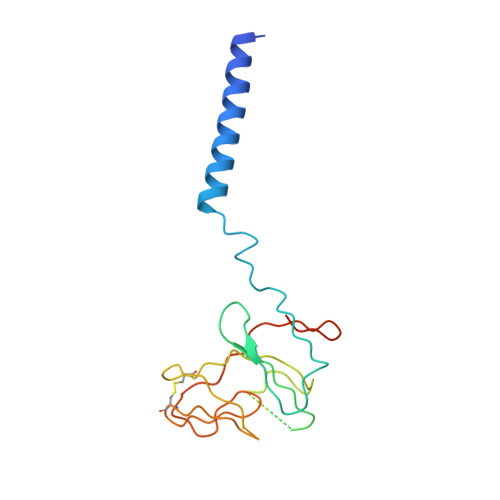
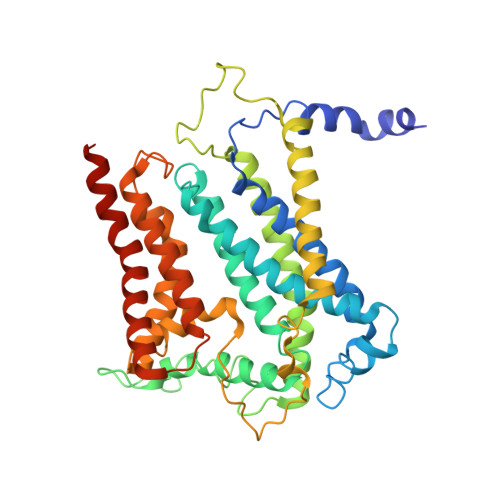
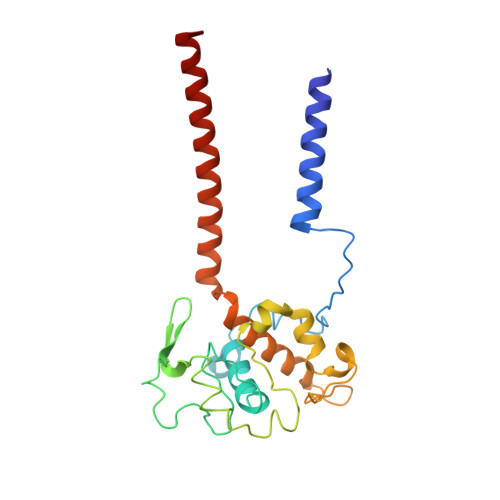
A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Zhu, G., Zeng, H., Zhang, S., Juli, J., Pang, X., Hoffmann, J., Zhang, Y., Morgner, N., Zhu, Y., Peng, G., Michel, H., Sun, F.(2020) Angew Chem Int Ed Engl 59: 343-351
- PubMed: 31778296
- DOI: https://doi.org/10.1002/anie.201911554
- Primary Citation of Related Structures:
6KLS, 6KLV - PubMed Abstract:
Respiratory chain complexes convert energy by coupling electron flow to transmembrane proton translocation. Owing to a lack of atomic structures of cytochrome bc 1 complex (Complex III) from thermophilic bacteria, little is known about the adaptations of this macromolecular machine to hyperthermophilic environments. In this study, we purified the cytochrome bc 1 complex of Aquifex aeolicus, one of the most extreme thermophilic bacteria known, and determined its structure with and without an inhibitor at 3.3 Å resolution. Several residues unique for thermophilic bacteria were detected that provide additional stabilization for the structure. An extra transmembrane helix at the N-terminus of cyt. c 1 was found to greatly enhance the interaction between cyt. b and cyt. c 1 , and to bind a phospholipid molecule to stabilize the complex in the membrane. These results provide the structural basis for the hyperstability of the cytochrome bc 1 complex in an extreme thermal environment.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics (IBP), Chinese Academy of Sciences, 15 Datun Road, Chaoyang District, Beijing, 100101, China.