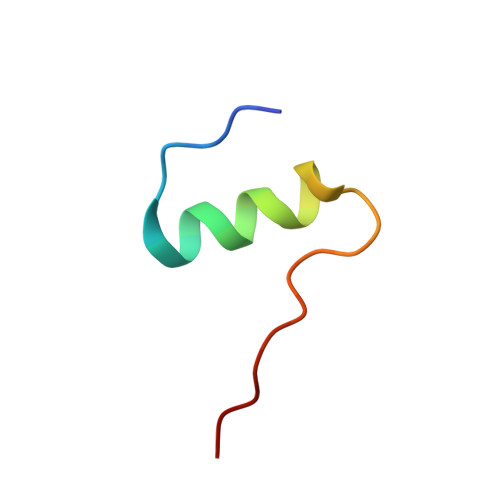
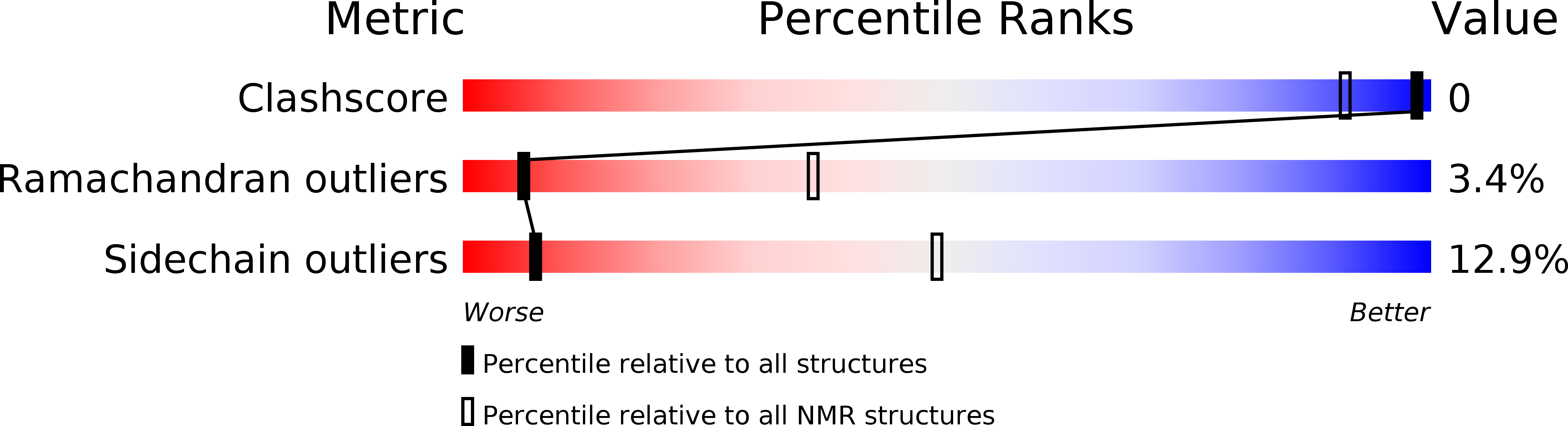High-resolution structure of a partially folded insulin aggregation intermediate.
Ratha, B.N., Kar, R.K., Brender, J.R., Pariary, R., Sahoo, B., Kalita, S., Bhunia, A.(2020) Proteins 88: 1648-1659
- PubMed: 32683793
- DOI: https://doi.org/10.1002/prot.25983
- Primary Citation of Related Structures:
6KH8, 6KH9, 6KHA - PubMed Abstract:
Insulin has long been served as a model for protein aggregation, both due to the importance of aggregation in the manufacture of insulin and because the structural biology of insulin has been extensively characterized. Despite intensive study, details about the initial triggers for aggregation have remained elusive at the molecular level. We show here that at acidic pH, the aggregation of insulin is likely initiated by a partially folded monomeric intermediate. High-resolution structures of the partially folded intermediate show that it is coarsely similar to the initial monomeric structure but differs in subtle details-the A chain helices on the receptor interface are more disordered and the B chain helix is displaced from the C-terminal A chain helix when compared to the stable monomer. The result of these movements is the creation of a hydrophobic cavity in the center of the protein that may serve as nucleation site for oligomer formation. Knowledge of this transition may aid in the engineering of insulin variants that retain the favorable pharamacokinetic properties of monomeric insulin but are more resistant to aggregation.
Organizational Affiliation:
Department of Biophysics, Bose Institute, Kolkata, India.