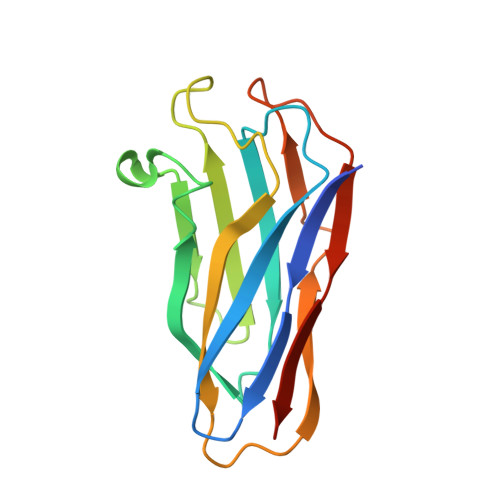
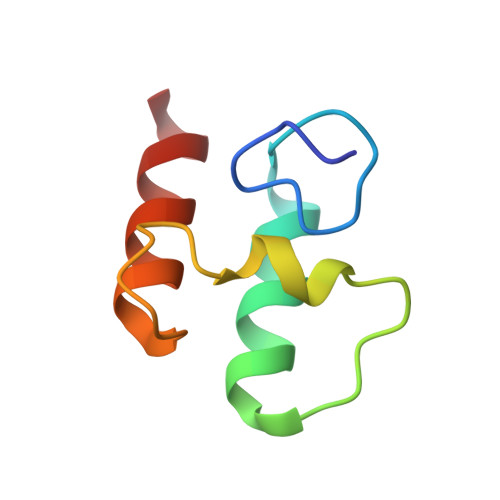
Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Yao, X., Chen, C., Wang, Y., Dong, S., Liu, Y.J., Li, Y., Cui, Z., Gong, W., Perrett, S., Yao, L., Lamed, R., Bayer, E.A., Cui, Q., Feng, Y.(2020) Sci Adv 6
- PubMed: 33097546
- DOI: https://doi.org/10.1126/sciadv.abd7182
- Primary Citation of Related Structures:
6KG8, 6KG9, 6KGC, 6KGD, 6KGE, 6KGF - PubMed Abstract:
Many important proteins undergo pH-dependent conformational changes resulting in "on-off" switches for protein function, which are essential for regulation of life processes and have wide application potential. Here, we report a pair of cellulosomal assembly modules, comprising a cohesin and a dockerin from Clostridium acetobutylicum , which interact together following a unique pH-dependent switch between two functional sites rather than on-off states. The two cohesin-binding sites on the dockerin are switched from one to the other at pH 4.8 and 7.5 with a 180° rotation of the bound dockerin. Combined analysis by nuclear magnetic resonance spectroscopy, crystal structure determination, mutagenesis, and isothermal titration calorimetry elucidates the chemical and structural mechanism of the pH-dependent switching of the binding sites. The pH-dependent dual-binding-site switch not only represents an elegant example of biological regulation but also provides a new approach for developing pH-dependent protein devices and biomaterials beyond an on-off switch for biotechnological applications.
Organizational Affiliation:
CAS Key Laboratory of Biofuels, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, Shandong 266101, China.