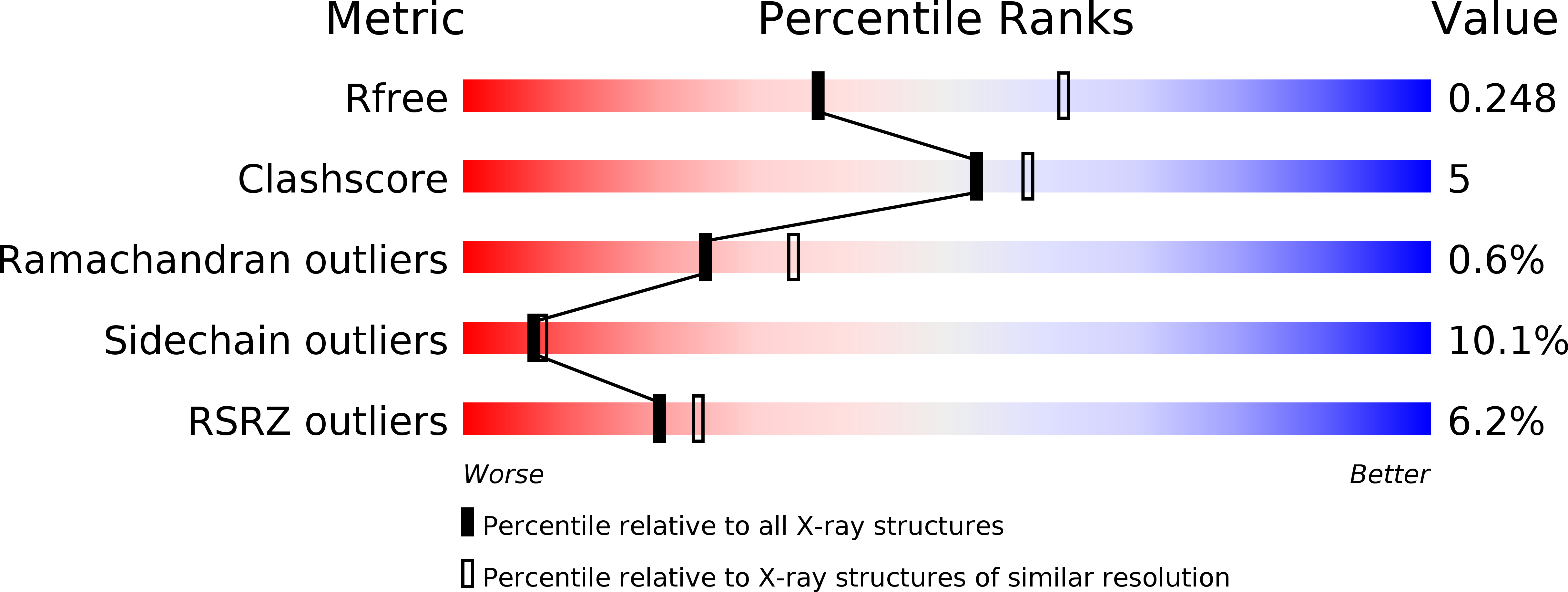
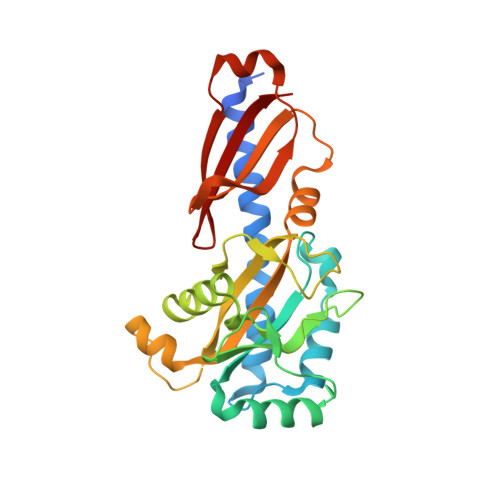
The crystal structure of the phytopathogenic bacterial sensor PcrK reveals different cytokinin recognition mechanism from the plant sensor AHK4.
Chen, P., Jiao, X., Zhang, Y., Wu, L., Tang, D.J., Li, P., Chen, X., Chao, D., Tang, J.L., Ming, Z.(2019) J Struct Biol 208: 69-76
- PubMed: 31419523
- DOI: https://doi.org/10.1016/j.jsb.2019.08.001
- Primary Citation of Related Structures:
6K62 - PubMed Abstract:
Plant cytokinins (CKs) are essential for many central cellular processes and play important roles in the interaction between bacteria and plants. Perception of CK is executed by the CHASE domain in the histidine kinase sensors of a class of two-component regulatory systems. Despite advances in understanding the structural basis for CK perception by the sensor AHK4 in Arabidopsis, the molecular mechanism of CK binding by other sensors is unclear. Here, we report the crystal structure of the CHASE domain in the histidine kinase PcrK of the bacterial plant pathogen Xanthomonas campestris pathovar campestris, which senses plant CK, determined at 2.55 Å resolution. The structure reveals that the PcrK has an AHK4-like overall topology and assembles into a homodimer. Strikingly, detailed structural analysis unveils two unique features of the PcrK ligand binding pocket: the size of the pocket is restricted for CK binding, and the PcrK applies a positively charged arginine but not a negatively charged aspartate to recognize the ligand. We propose a model to explain how the PcrK accommodates CK-sized compounds through conformational changes, providing a potential mechanistic framework for understanding ligand recognition by the PcrK.
Organizational Affiliation:
State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, 100 Daxue Road, Nanning, Guangxi 530004, China.