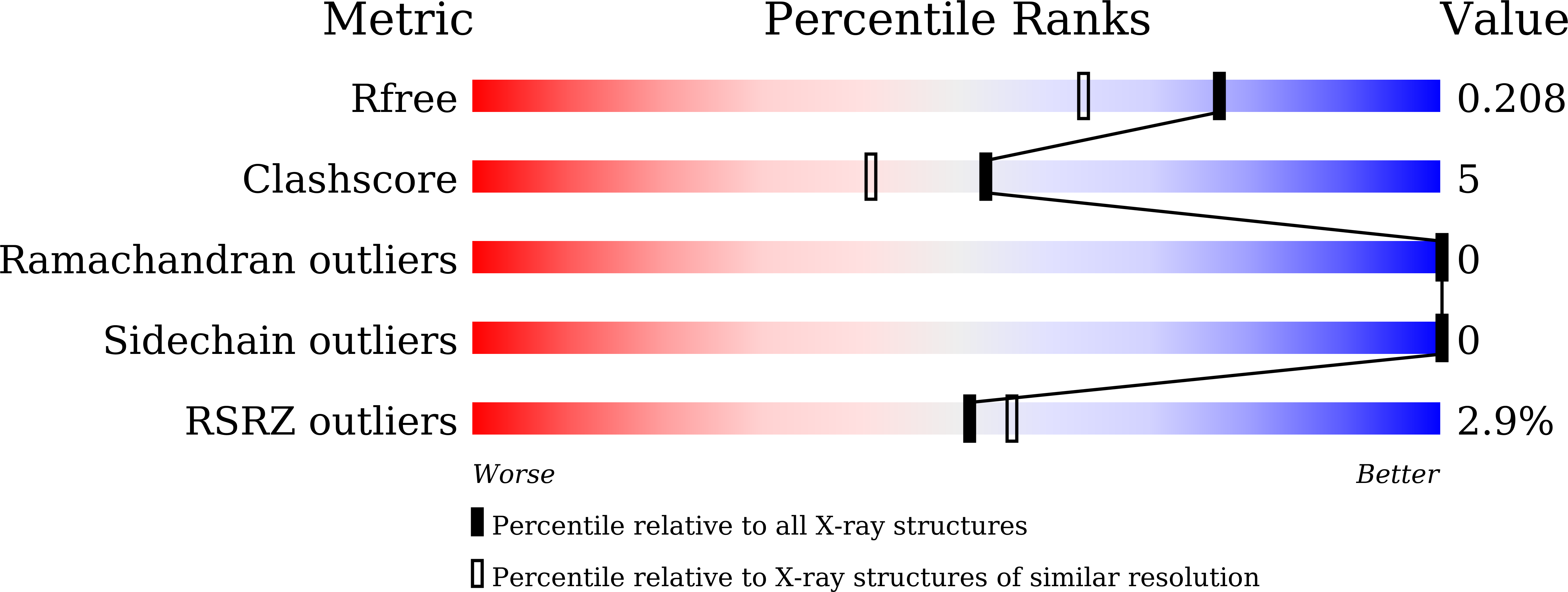
Structural Basis of Leader Peptide Recognition in Lasso Peptide Biosynthesis Pathway.
Sumida, T., Dubiley, S., Wilcox, B., Severinov, K., Tagami, S.(2019) ACS Chem Biol 14: 1619-1627
- PubMed: 31188556
- DOI: https://doi.org/10.1021/acschembio.9b00348
- Primary Citation of Related Structures:
6JX3 - PubMed Abstract:
Lasso peptides are a class of ribosomally synthesized and post-translationally modified peptides (RiPPs) with a unique 3D-interlocked structure, in which an N-terminal macrolactam ring is threaded by a linear C-terminal part. The unique structure of lasso peptides is introduced into ribosomally translated precursor peptides by lasso peptide synthetase encompassing proteins B and C or B1, B2, and C when the B enzyme is split into two distinct proteins. The B1 protein recognizes the leader sequence of the precursor peptide, and then the B2 protein cleaves it. The C protein catalyzes the formation of the macrolactam ring. However, the detailed mechanism of lasso peptide maturation has remained elusive, due to the lack of structural information about the responsible proteins. Here we report the crystal structure of the B1 protein from the thermophilic actinobacteria, Thermobifida fusca (TfuB1), complexed with the leader peptide (TfuA-Leader), which revealed the detailed mechanism of leader peptide recognition. The structure of TfuB1 consists of an N-terminal β-sheet and three C-terminal helices. The leader peptide is docked on one edge of the N-terminal β-sheet of TfuB1, as an additional β strand. Three conserved amino acid residues of the leader peptide (TfuA Tyr-17, Pro-14, and Leu-12) fit well on the hydrophobic cleft between the β-sheet and adjacent helices. Biochemical analysis demonstrated that these conserved residues are essential for affinity between TfuB1 and the TfuA-Leader. Furthermore, we found that TfuB1 and the leader peptide jointly form a hydrophobic patch on the β-sheet, which includes the highly conserved TfuA Phe-6 and TfuB1 Tyr33. Homology modeling and mutational analysis of the B1 protein from a firmicute, Bacillus pseudomycoides (PsmB1), revealed that the hydrophobic patch is conserved in a wide range of species and involved in the cleavage activity of the B2 protein, indicating it forms the interaction surface for the B2 protein or the core part of the precursor peptide.
Organizational Affiliation:
Research Center for Bioscience and Nanoscience , Japan Agency for Marine-Earth Science and Technology (JAMSTEC) , 2-15 Natsushima-cho , Yokosuka 237-0061 , Japan.