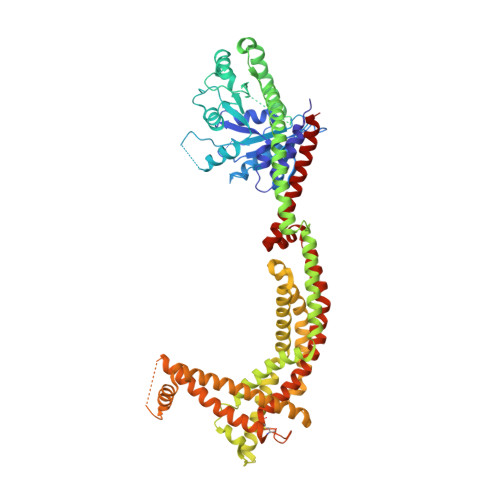
Structural analysis of a trimeric assembly of the mitochondrial dynamin-like GTPase Mgm1.
Yan, L., Qi, Y., Ricketson, D., Li, L., Subramanian, K., Zhao, J., Yu, C., Wu, L., Sarsam, R., Wong, M., Lou, Z., Rao, Z., Nunnari, J., Hu, J.(2020) Proc Natl Acad Sci U S A 117: 4061-4070
- PubMed: 32041880
- DOI: https://doi.org/10.1073/pnas.1919116117
- Primary Citation of Related Structures:
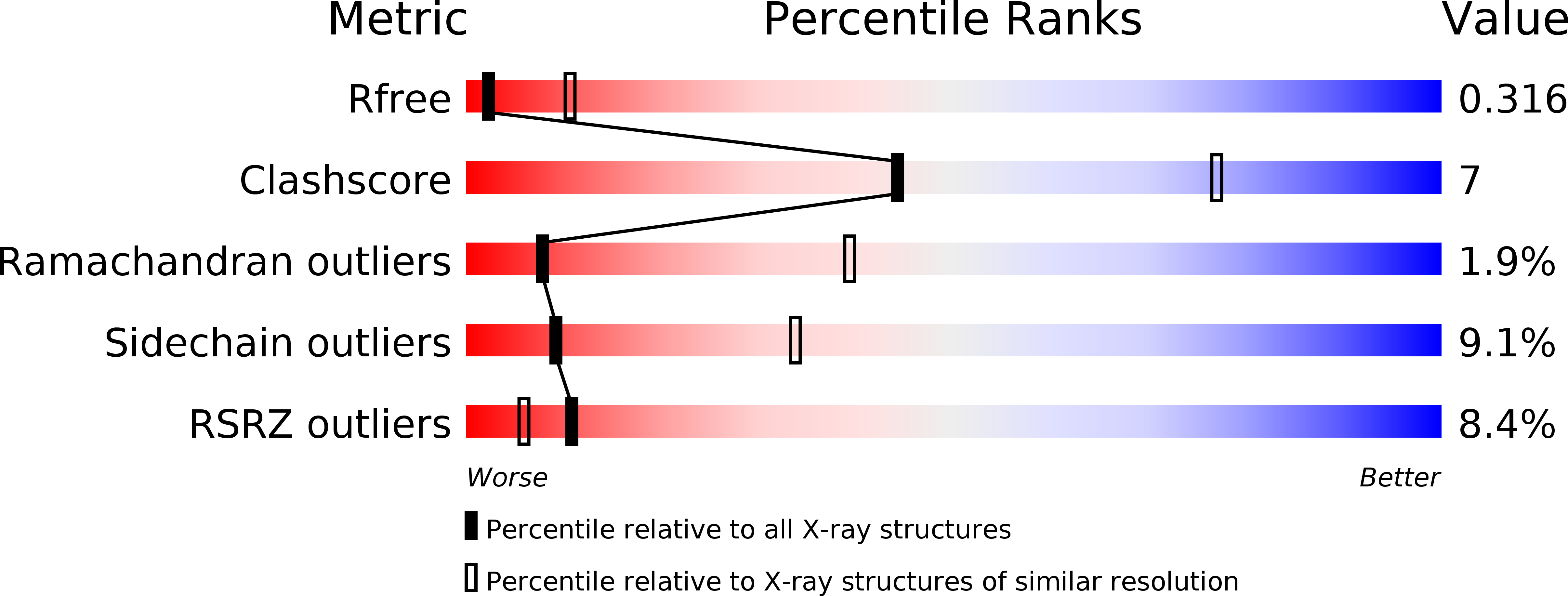
6JSJ - PubMed Abstract:
The fusion of inner mitochondrial membranes requires dynamin-like GTPases, Mgm1 in yeast and OPA1 in mammals, but how they mediate membrane fusion is poorly understood. Here, we determined the crystal structure of Saccharomyces cerevisiae short Mgm1 (s-Mgm1) in complex with GDP. It revealed an N-terminal GTPase (G) domain followed by two helix bundles (HB1 and HB2) and a unique C-terminal lipid-interacting stalk (LIS). Dimers can form through antiparallel HB interactions. Head-to-tail trimers are built by intermolecular interactions between the G domain and HB2-LIS. Biochemical and in vivo analyses support the idea that the assembly interfaces observed here are native and critical for Mgm1 function. We also found that s-Mgm1 interacts with negatively charged lipids via both the G domain and LIS. Based on these observations, we propose that membrane targeting via the G domain and LIS facilitates the in cis assembly of Mgm1, potentially generating a highly curved membrane tip to allow inner membrane fusion.
Organizational Affiliation:
National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.