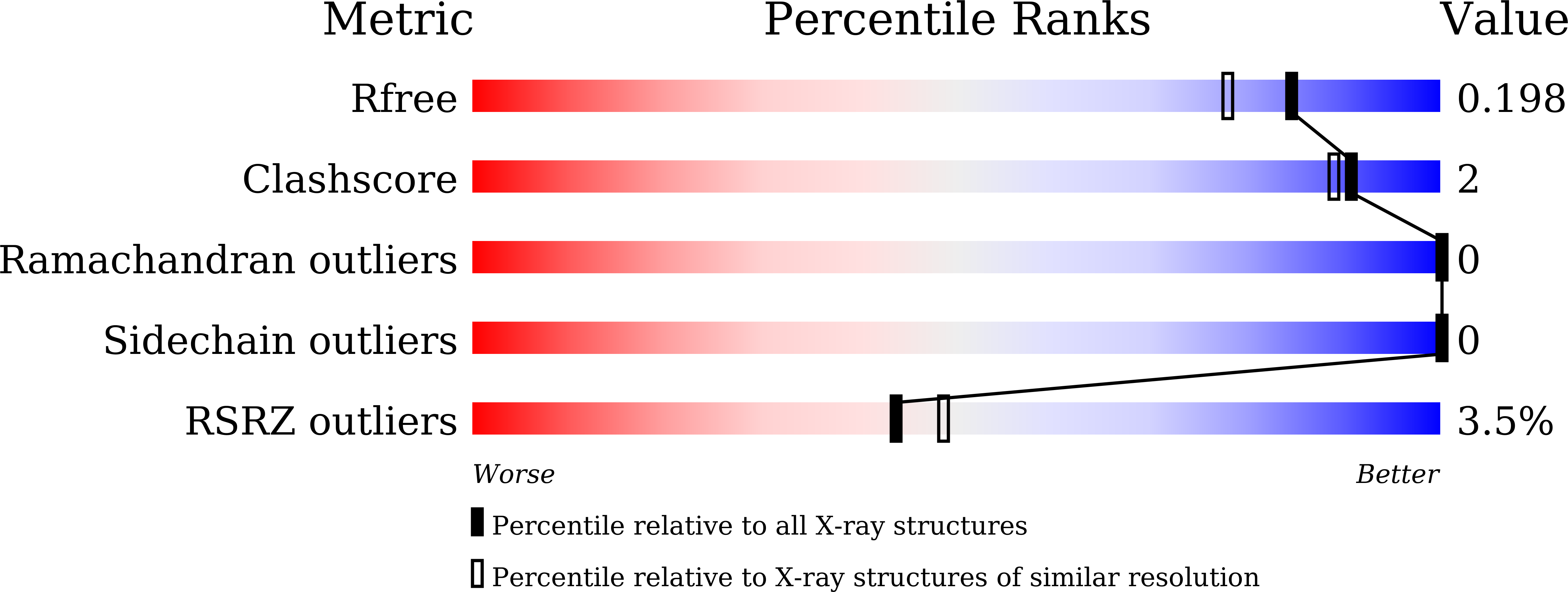
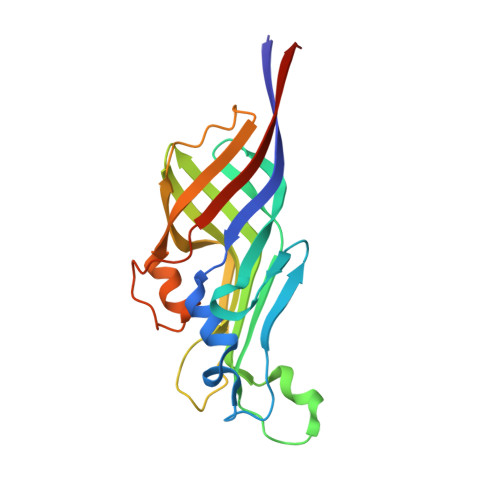
Structural basis for the heme transfer reaction in heme uptake machinery from Corynebacteria.
Muraki, N., Kitatsuji, C., Okamoto, Y., Uchida, T., Ishimori, K., Aono, S.(2019) Chem Commun (Camb) 55: 13864-13867
- PubMed: 31670736
- DOI: https://doi.org/10.1039/c9cc07369h
- Primary Citation of Related Structures:
6JS9, 6JSA, 6JSB, 6JSC, 6JSD - PubMed Abstract:
The crystal structures of the conserved region domains of HtaA and HtaB, which act as heme binding/transport proteins in the heme uptake machinery in Corynebacterium glutamicum, are determined for the first time. The molecular mechanism of heme transfer among these proteins is proposed based on the spectroscopic and structural analyses.
Organizational Affiliation:
Department of Creative Research, Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences, 5-1 Higashiyama, Myodaiji-cho, Okazaki 444-8787, Japan. aono@ims.ac.jp.