Crystal structure of phyllogen, a phyllody-inducing effector protein of phytoplasma.
Iwabuchi, N., Maejima, K., Kitazawa, Y., Miyatake, H., Nishikawa, M., Tokuda, R., Koinuma, H., Miyazaki, A., Nijo, T., Oshima, K., Yamaji, Y., Namba, S.(2019) Biochem Biophys Res Commun 513: 952-957
- PubMed: 31010685
- DOI: https://doi.org/10.1016/j.bbrc.2019.04.060
- Primary Citation of Related Structures:
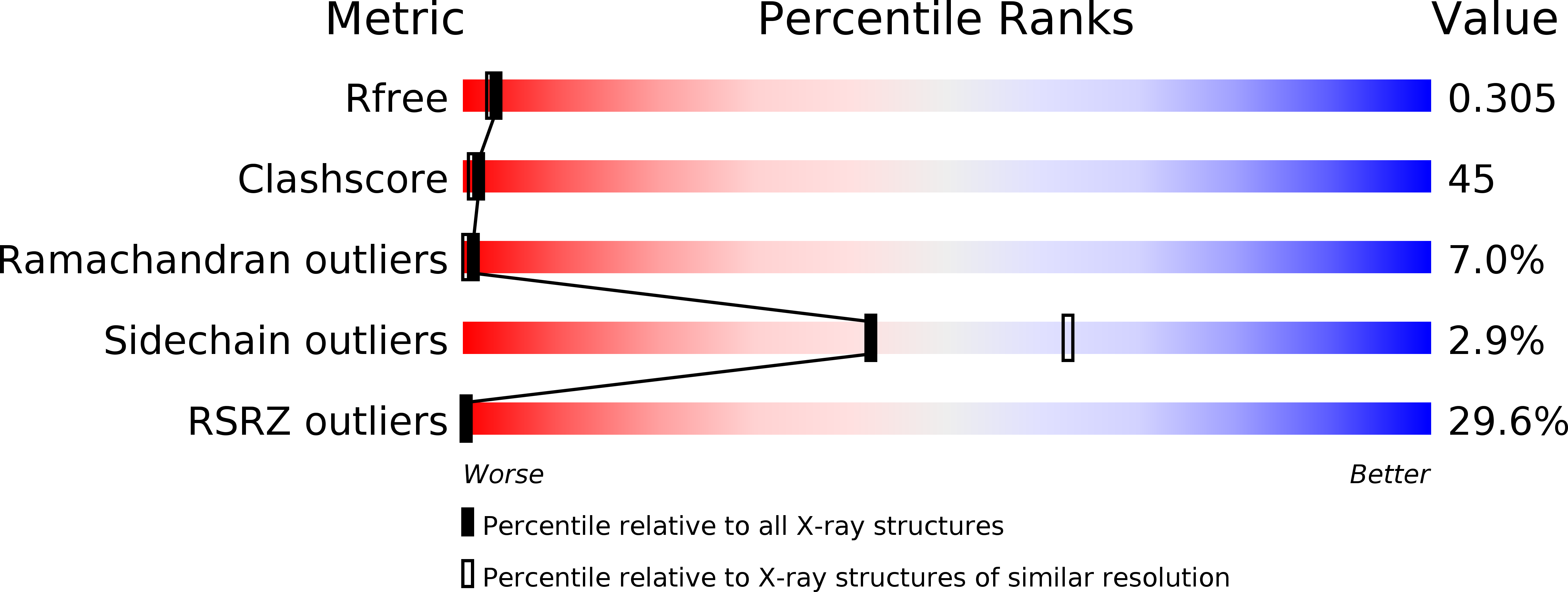
6JQA - PubMed Abstract:
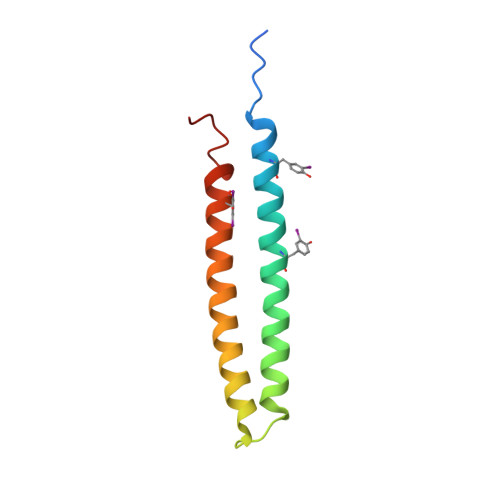
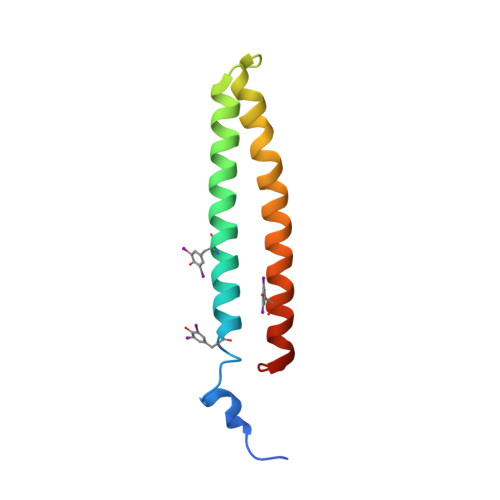
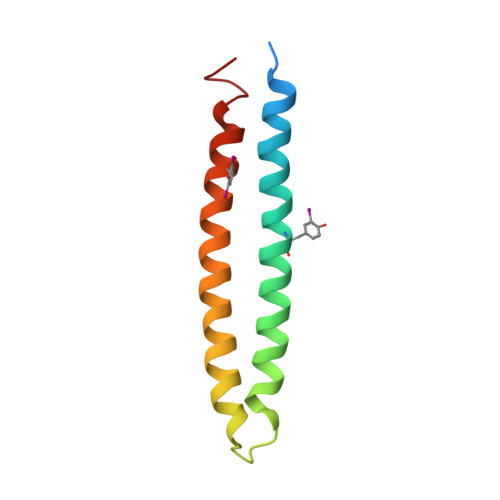
Phytoplasmas are plant pathogenic bacteria that often induce unique phyllody symptoms in which the floral organs are transformed into leaf-like structures. Recently, a novel family of bacterial effector genes, called phyllody-inducing genes (phyllogens), was identified as being involved in the induction of phyllody by degrading floral MADS-domain transcription factors (MTFs). However, the structural characteristics of phyllogens are unknown. In this study, we elucidated the crystal structure of PHYL1 OY , a phyllogen of 'Candidatus Phytoplasma asteris' onion yellows strain, at a resolution of 2.4 Å. The structure of PHYL1 consisted of two α-helices connected by a random loop in a coiled-coil manner. In both α-helices, the distributions of hydrophobic residues were conserved among phyllogens. Amino acid insertion mutations into either α-helix resulted in the loss of phyllody-inducing activity and the ability of the phyllogen to degrade floral MTF. In contrast, the same insertion in the loop region did not affect either activity, indicating that both conserved α-helices are important for the function of phyllogens. This is the first report on the crystal structure of an effector protein of phytoplasmas.
Organizational Affiliation:
Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, 113-8657, Japan.