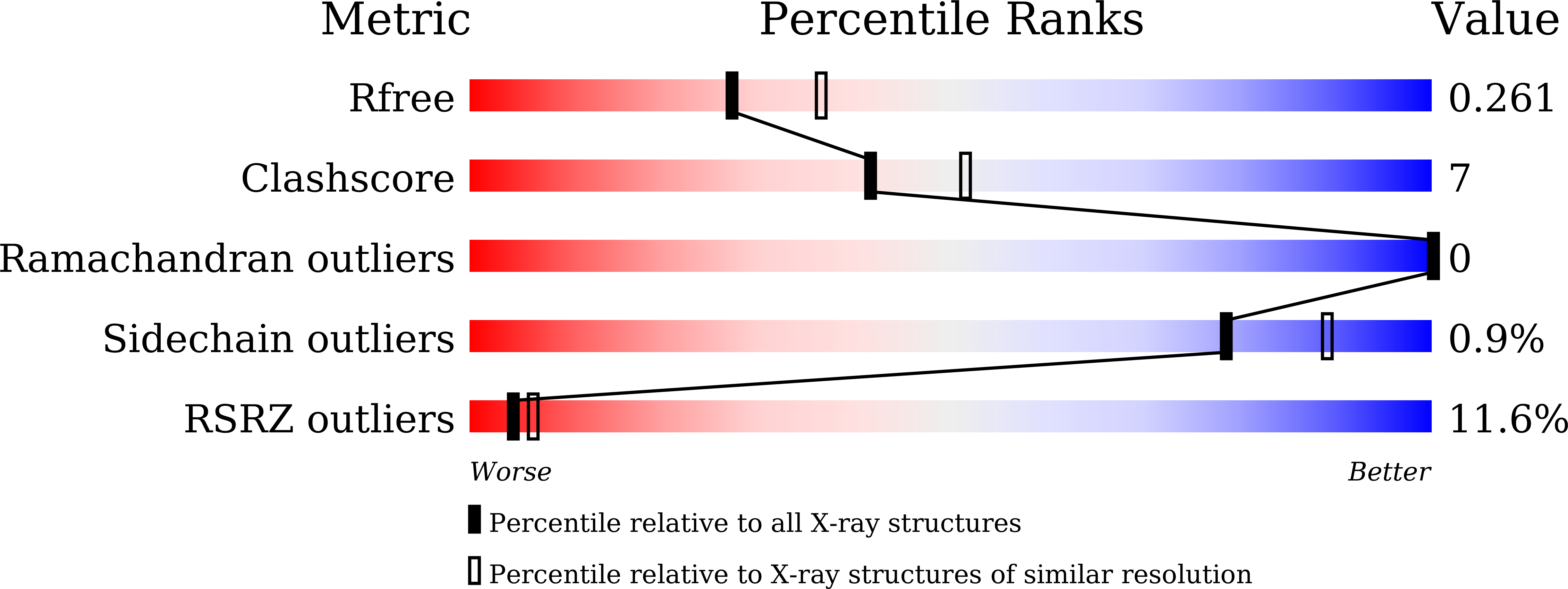
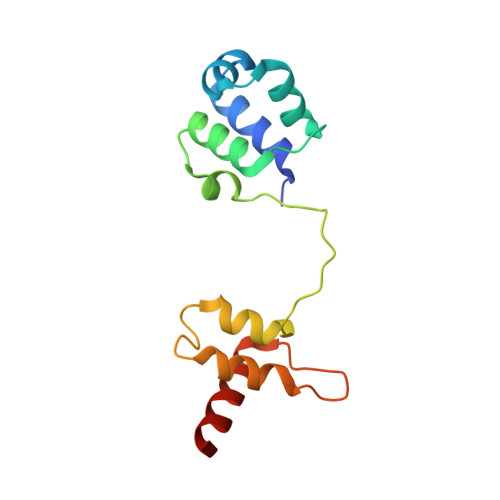
Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO.
Lu, H., Wang, L., Li, S., Pan, C., Cheng, K., Luo, Y., Xu, H., Tian, B., Zhao, Y., Hua, Y.(2019) Nucleic Acids Res 47: 9925-9933
- PubMed: 31410466
- DOI: https://doi.org/10.1093/nar/gkz720
- Primary Citation of Related Structures:
6JQ1 - PubMed Abstract:
DdrO is an XRE family transcription repressor that, in coordination with the metalloprotease PprI, is critical in the DNA damage response of Deinococcus species. Here, we report the crystal structure of Deinococcus geothermalis DdrO. Biochemical and structural studies revealed the conserved recognizing α-helix and extended dimeric interaction of the DdrO protein, which are essential for promoter DNA binding. Two conserved oppositely charged residues in the HTH motif of XRE family proteins form salt bridge interactions that are essential for promoter DNA binding. Notably, the C-terminal domain is stabilized by hydrophobic interactions of leucine/isoleucine-rich helices, which is critical for DdrO dimerization. Our findings suggest that DdrO is a novel XRE family transcriptional regulator that forms a distinctive dimer. The structure also provides insight into the mechanism of DdrO-PprI-mediated DNA damage response in Deinococcus.
Organizational Affiliation:
MOE Key Laboratory of Biosystems Homeostasis & Protection, Zhejiang University, China.