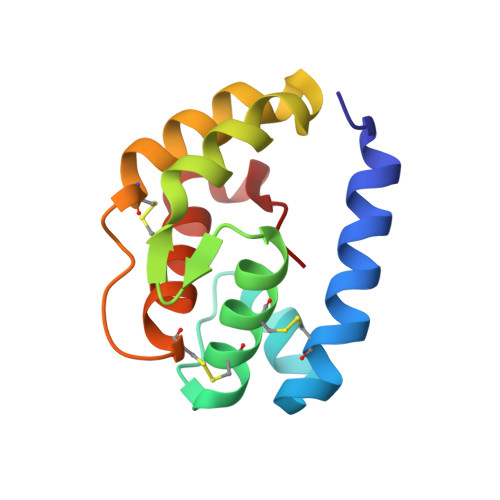
Crystal structure and ligand identification of odorant binding protein 4 in the natural predator Chrysopa pallens.
Li, T.T., Liu, W.C., Zhu, J., Yang, Y.H., Ma, C., Lu, C., Zhang, K.X.(2019) Int J Biol Macromol 141: 1004-1012
- PubMed: 31525411
- DOI: https://doi.org/10.1016/j.ijbiomac.2019.09.043
- Primary Citation of Related Structures:
6JPM - PubMed Abstract:
Green lacewing Chrysopa pallens (Rambur) is a general predator of many agricultural pests and plays a pivotal role in reducing crop damage by managing insect pest populations. Odorant binding proteins (OBPs) in insects can sense the semiochemicals in the environment and initiate the delivery of signals to their receptors. However, no Chrysopa pallens OBP (CpalOBP) structure has been reported yet, and their corresponding candidate semiochemicals are still largely unknown. Here, we reported the structure of CpalOBP4 solved with X-ray diffraction and showed its potential ligands. Our results showed that CpalOBP4 has a classical OBP structure with six α-helices and three disulfide bridges, and it can bind with farnesene, 2-tridecanone, cis-3-hexenyl hexanoate, nerolidol and farnesol through a central hydrophobic cavity. Our molecular docking results showed that Met31, Met78, Leu98, Phe141, Leu142 and Pro143 in the hydrophobic cavity were the key residues mediating the interaction of CpalOBP4 with farnesene, 2-tridecanone and cis-3-hexenyl hexanoate, which was further proven by the results that mutations of these residues led to significantly reduced binding affinities of CpalOBP4 for these ligands. Our study provides useful information for the further investigation of the biological function of CpalOBP4 as well as important cues for improving biological control in agriculture.
Organizational Affiliation:
Jiangsu Key Laboratory of Marine Biological Resources and Environment, Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening, Huaihai Institute of Technology, Lianyungang 222005, China; Co-Innovation Center of Jiangsu Marine Bio-industry Technology, Huaihai Institute of Technology, Lianyungang 222005, China.