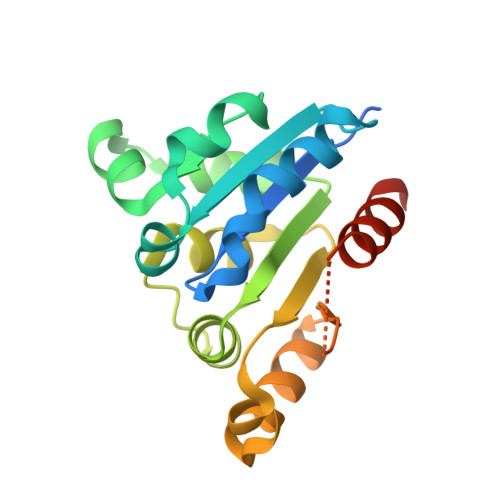
Characterization of the FMN-Dependent Cysteine Decarboxylase from Thioviridamide Biosynthesis.
Lu, J., Li, J., Wu, Y., Fang, X., Zhu, J., Wang, H.(2019) Org Lett 21: 4676-4679
- PubMed: 31184189
- DOI: https://doi.org/10.1021/acs.orglett.9b01531
- Primary Citation of Related Structures:
6JLS - PubMed Abstract:
The biosynthesis of thioviridamide-like compounds has not been elucidated. Herein, we report that TvaF from the thioviridamide biosynthetic gene cluster is an FMN-dependent cysteine decarboxylase that transforms the C-terminal cysteine of precursor peptides into a thioenol motif and exhibits high substrate flexibility. We resolved the crystal structure of TvaF bound with FMN at 2.24 Å resolution. Key residues for FMN binding and catalytic activity of TvaF have been identified and evaluated by mutagenesis studies.
Organizational Affiliation:
State Key Laboratory of Coordination Chemistry, Jiangsu Key Laboratory of Advanced Organic Materials, School of Chemistry and Chemical Engineering , Nanjing University , Nanjing 210023 , China.