Structural insight into endonuclease CRISPR-associated Cas2 protein
Bi, M., Mo, X., Wang, C., Yuan, A.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CRISPR-associated endoribonuclease Cas2 | 93 | Treponema denticola ATCC 35405 | Mutation(s): 0 Gene Names: cas2, TDE_0329 EC: 3.1 | 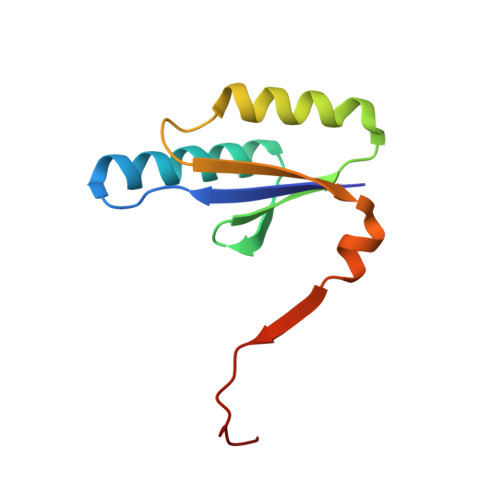 | |
UniProt | |||||
Find proteins for Q73QW4 (Treponema denticola (strain ATCC 35405 / DSM 14222 / CIP 103919 / JCM 8153 / KCTC 15104)) Explore Q73QW4 Go to UniProtKB: Q73QW4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q73QW4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5-mer peptide | C [auth Q] | 5 | Escherichia coli | Mutation(s): 0 |  |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 26.592 | α = 90 |
| b = 76.391 | β = 90 |
| c = 88.445 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MoRDa | phasing |