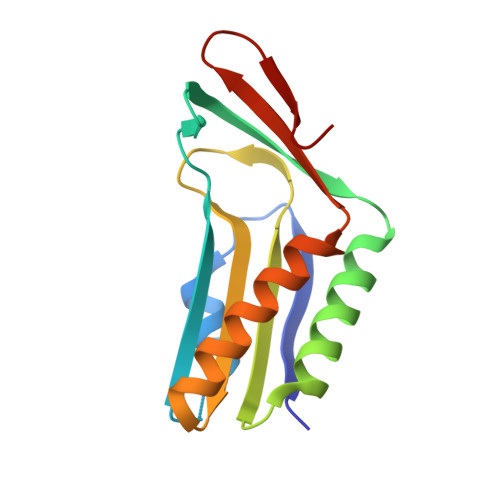
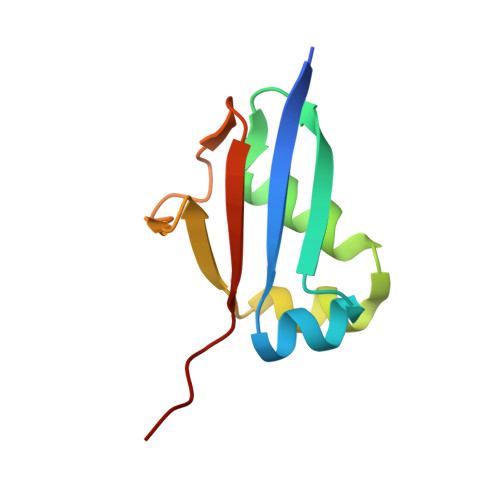
Structural analysis of molybdopterin synthases from two mycobacterial pathogens.
Wang, H., Chen, X., Zhang, W., Zhou, W., Liu, X., Rao, Z.(2019) Biochem Biophys Res Commun 511: 21-27
- PubMed: 30765225
- DOI: https://doi.org/10.1016/j.bbrc.2019.02.024
- Primary Citation of Related Structures:
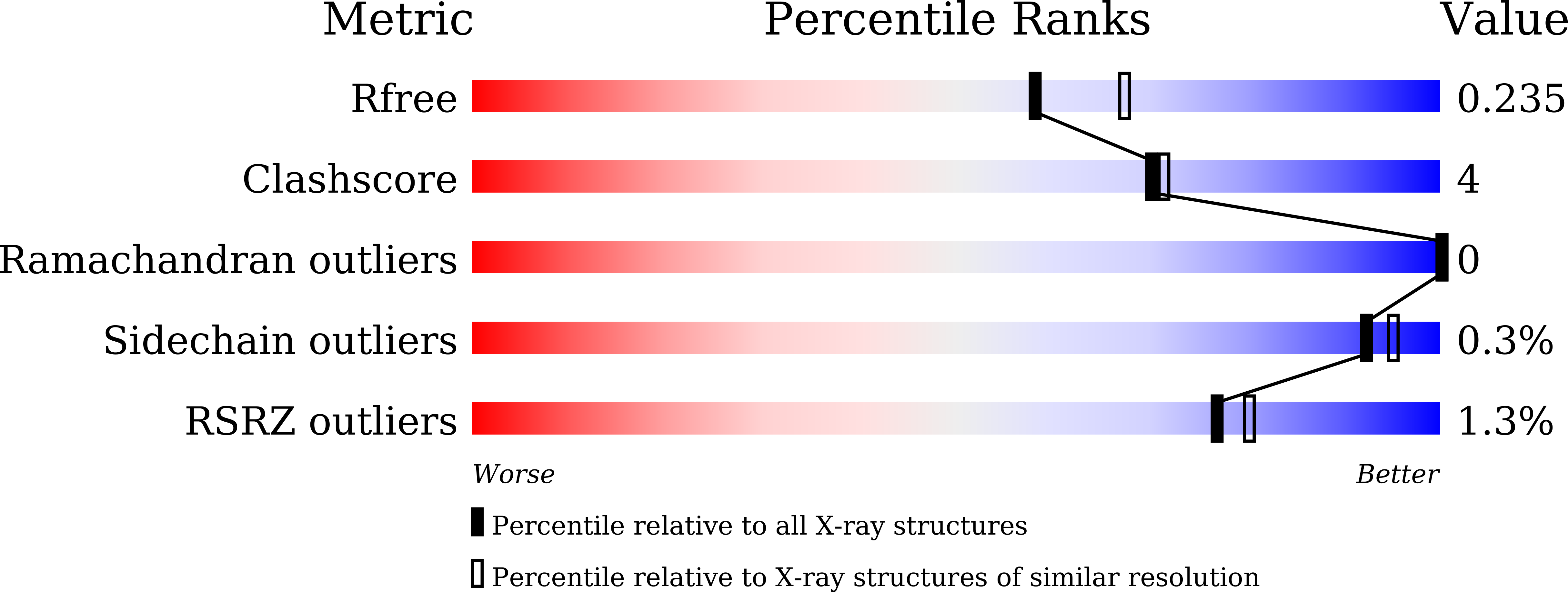
6JBZ, 6JC0 - PubMed Abstract:
The molybdenum cofactor, composed of molybdopterin and molybdenum, is a necessary compound for the catalytic activity of molybdenum enzymes. Molybdenum cofactor biosynthesis is a conserved multi-step process involving several enzymes. Molybdopterin synthase, a hetero-tetrameric enzyme composed of a pair of MoaE-MoaD subunits, catalyzes the generation of the cis-dithiolene group of molybdopterin in the second step of the process. The cis-dithiolene group can covalently bind molybdenum. Most mycobacterial species possess several genes encoding the full pathway of molybdenum cofactor biosynthesis. In M. smegmatis, the moaD2 and moaE2 genes encode the functional molybdopterin synthase. However, M. tuberculosis has genes encoding several molybdopterin synthase subunit homologs, including moaD1, moaD2, moaE1, moaE2, and moaX, which encodes a MoaD-MoaE fusion protein. Previous studies have shown that moaD2 and moaE2 encode functional molybdopterin synthase. Here, we report the crystal structures of two substrate-free molybdopterin synthases from two different mycobacterial pathogens, M. tuberculosis and M. smegmatis, at 2.1 Å and 2.6 Å resolutions, respectively. The overall structure of both molybdopterin synthases was hetero-tetrameric, consisting of a MoaE2 dimer flanked on either side by single MoaD2 subunits. The carboxyl-terminal domain of MoaD2 inserted into MoaE2, forming the active pocket. A comparison with previously reported molybdopterin synthase structures showed that substrate-binding and catalytic residues were conserved, despite low sequence similarity among these enzymes. The low sequence identity at the MoaE-MoaD heterodimer interface may provide the structural basis to explore mycobacterial inhibitors.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology and College of Pharmacy, Nankai University, Tianjin, China; College of Life Science, Nankai University, Tianjin, China.