Phytophthora sojae Effector PsAvh240 Inhibits Host Aspartic Protease Secretion to Promote Infection.
Guo, B., Wang, H., Yang, B., Jiang, W., Jing, M., Li, H., Xia, Y., Xu, Y., Hu, Q., Wang, F., Yu, F., Wang, Y., Ye, W., Dong, S., Xing, W., Wang, Y.(2019) Mol Plant 12: 552-564
- PubMed: 30703565
- DOI: https://doi.org/10.1016/j.molp.2019.01.017
- Primary Citation of Related Structures:
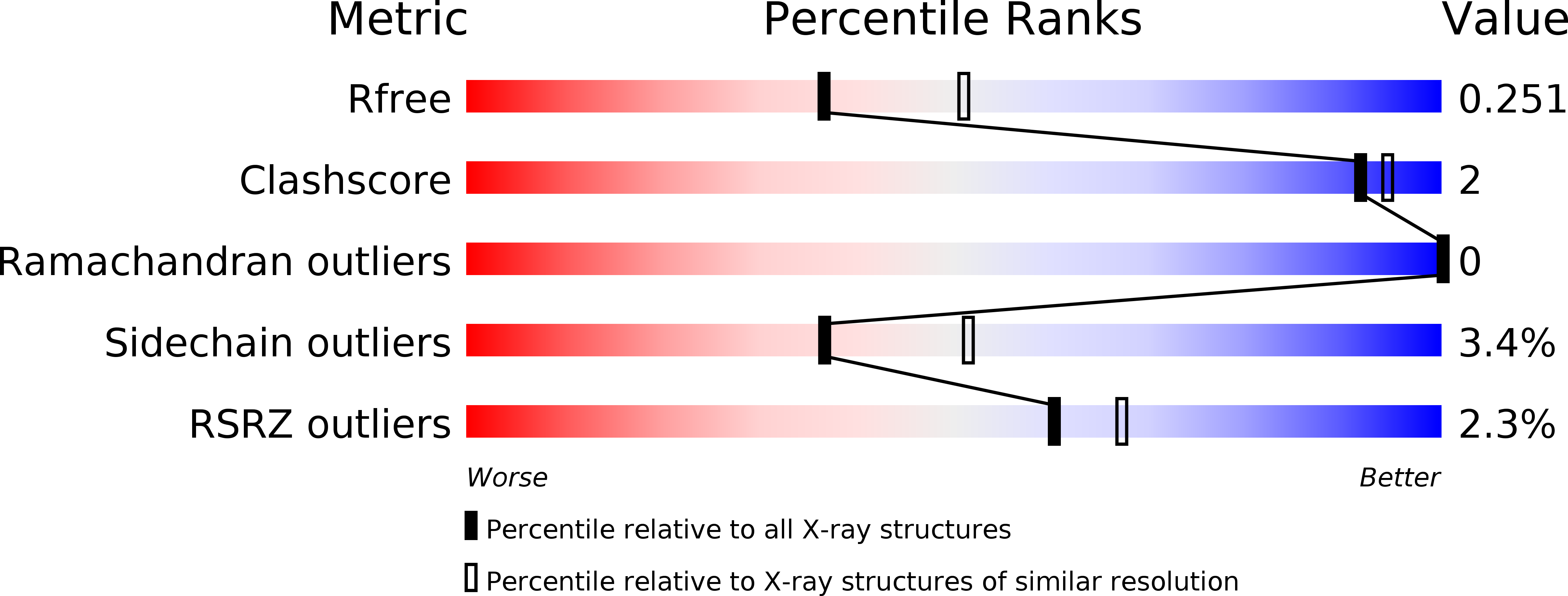
6J8L - PubMed Abstract:
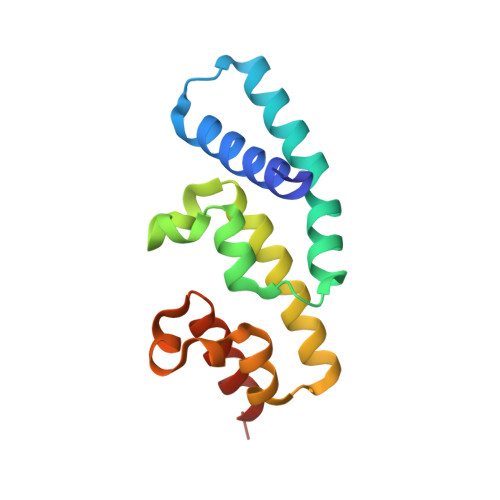
Plants secrete defense molecules into the extracellular space (the apoplast) to combat attacking microbes. However, the mechanisms by which successful pathogens subvert plant apoplastic immunity remain poorly understood. In this study, we show that PsAvh240, a membrane-localized effector of the soybean pathogen Phytophthora sojae, promotes P. sojae infection in soybean hairy roots. We found that PsAvh240 interacts with the soybean-resistant aspartic protease GmAP1 in planta and suppresses the secretion of GmAP1 into the apoplast. By solving its crystal structure we revealed that PsAvh240 contain six α helices and two WY motifs. The first two α helices of PsAvh240 are responsible for its plasma membrane-localization and are required for PsAvh240's interaction with GmAP1. The second WY motifs of two PsAvh240 molecules form a handshake arrangement resulting in a handshake-like dimer. This dimerization is required for the effector's repression of GmAP1 secretion. Taken together, these data reveal that PsAvh240 localizes at the plasma membrane to interfere with GmAP1 secretion, which represents an effective mechanism by which effector proteins suppress plant apoplastic immunity.
Organizational Affiliation:
Department of Plant Pathology, Nanjing Agricultural University, Nanjing 210095, China; Key Laboratory of Integrated Management of Crop Diseases and Pests (Ministry of Education), Nanjing 210095, China.