Crystal structure of the flavin-dependent thymidylate synthase Thy1 from Thermus thermophilus with an extra C-terminal domain.
Ogawa, A., Sampei, G., Kawai, G.(2019) Acta Crystallogr F Struct Biol Commun 75: 450-454
- PubMed: 31204692
- DOI: https://doi.org/10.1107/S2053230X19007192
- Primary Citation of Related Structures:
6J61 - PubMed Abstract:
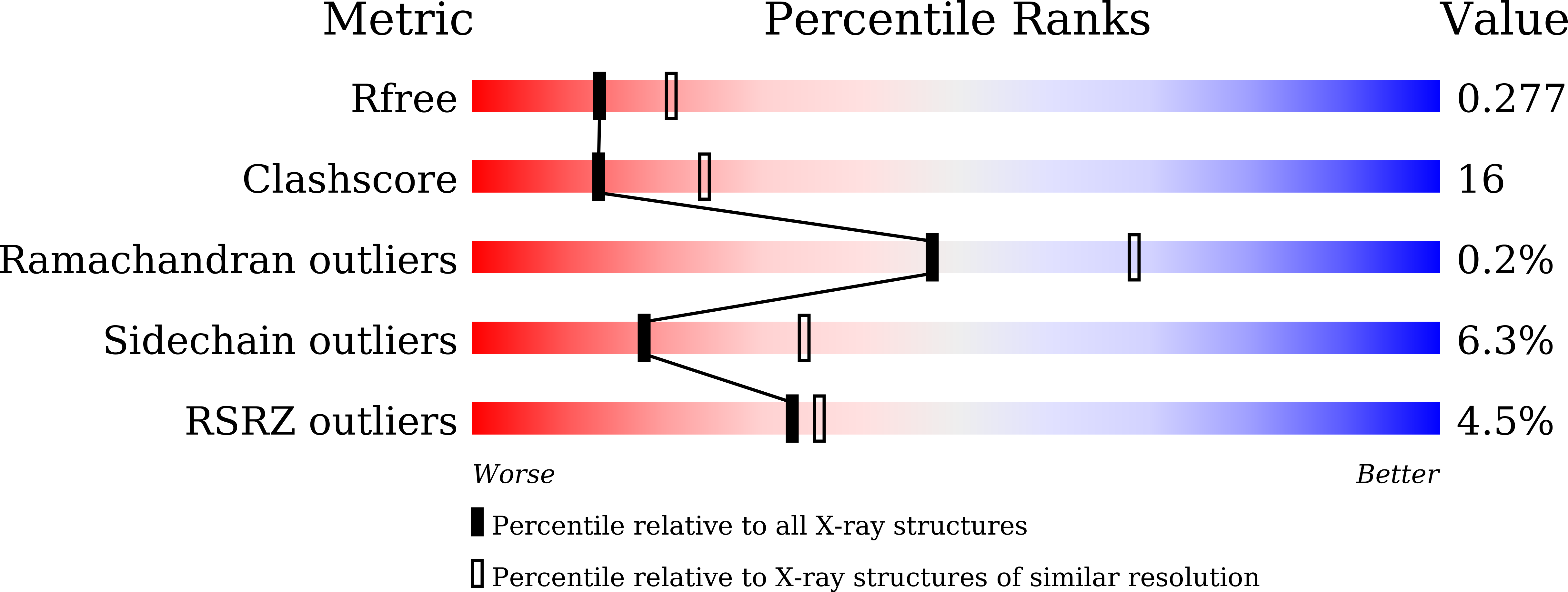
The thymidylate synthases ThyA and Thy1 are enzymes that catalyse the formation of thymidine monophosphate from 2'-deoxyuridine monophosphate. Thy1 (or ThyX) requires flavin for catalytic reactions, while ThyA does not. In the present study, the crystal structure of the flavin-dependent thymidylate synthase Thy1 from Thermus thermophilus HB8 (TtThy1, TTHA1096) was determined in complex with FAD and phosphate at 2.5 Å resolution. TtThy1 is a tetrameric molecule like other Thy1 proteins, to which four FAD molecules are bound. In the crystal of TtThy1, two phosphate ions were bound to each dUMP-binding site. The characteristic feature of TtThy1 is the existence of an extra C-terminal domain (CTD) consisting of three α-helices and a β-strand. The function of the CTD is unknown and database analysis showed that this CTD is only shared by part of the Deinococcus-Thermus phylum.
Organizational Affiliation:
Department of Life and Environmental Sciences, Faculty of Engineering, Chiba Institute of Technology, Narashino, Chiba 275-0016, Japan.