The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Park, K.S., Hong, M.K., Jeon, J.W., Kim, J.H., Jeon, J.H., Lee, J.H., Kim, T.Y., Karim, A.M., Malik, S.K., Kang, L.W., Lee, S.H.(2018) Acta Crystallogr F Struct Biol Commun 74: 644-649
- PubMed: 30279316
- DOI: https://doi.org/10.1107/S2053230X18012268
- Primary Citation of Related Structures:
6J4N - PubMed Abstract:
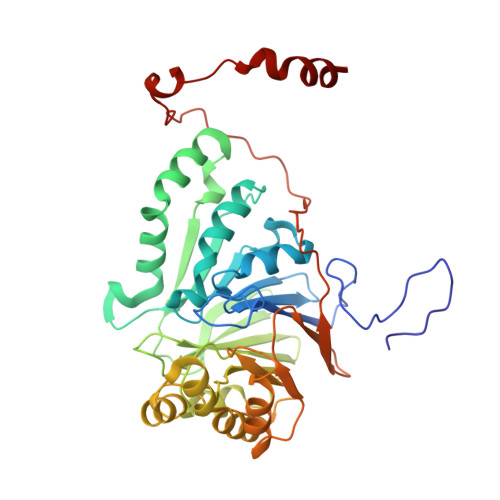
Metallo-β-lactamases (MBLs) are present in major Gram-negative pathogens and environmental species, and pose great health risks because of their ability to hydrolyze the β-lactam rings of antibiotics such as carbapenems. PNGM-1 was the first reported case of a subclass B3 MBL protein that was identified from a metagenomic library from deep-sea sediments that predate the antibiotic era. In this study, PNGM-1 was overexpressed, purified and crystallized. Crystals of native and selenomethionine-substituted PNGM-1 diffracted to 2.10 and 2.30 Å resolution, respectively. Both the native and the selenomethionine-labelled PNGM-1 crystals belonged to the monoclinic space group P2 1 , with unit-cell parameters a = 122, b = 83, c = 163 Å, β = 110°. Matthews coefficient (V M ) calculations suggested the presence of 6-10 molecules in the asymmetric unit, corresponding to a solvent content of ∼31-58%. Structure determination is currently in progress.
Organizational Affiliation:
National Leading Research Laboratory of Drug Resistance Proteomics, Department of Biological Sciences, Myongji University, 116 Myongjiro, Yongin, Gyeonggido 17058, Republic of Korea.