Structural and functional characterization of a bifunctional GH30-7 xylanase B from the filamentous fungusTalaromyces cellulolyticus.
Nakamichi, Y., Fouquet, T., Ito, S., Watanabe, M., Matsushika, A., Inoue, H.(2019) J Biol Chem 294: 4065-4078
- PubMed: 30655295
- DOI: https://doi.org/10.1074/jbc.RA118.007207
- Primary Citation of Related Structures:
6IUJ - PubMed Abstract:
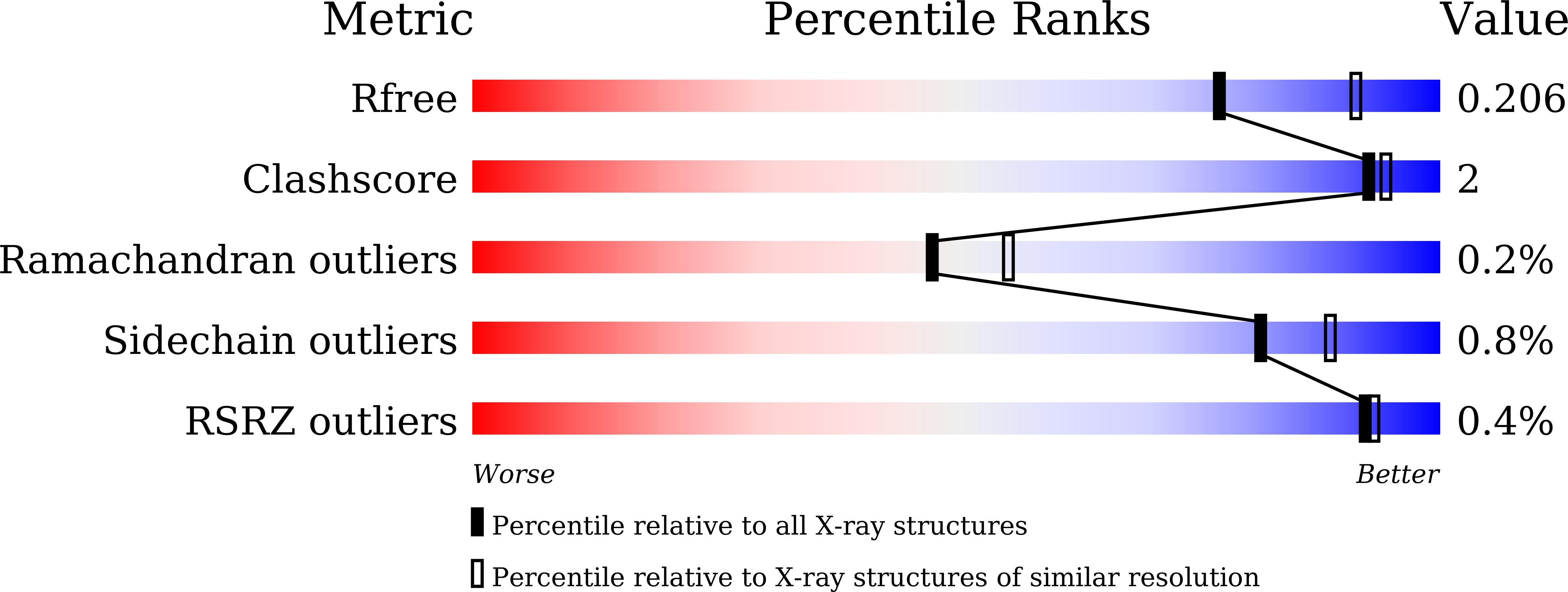
Glucuronoxylanases are endo-xylanases and members of the glycoside hydrolase family 30 subfamilies 7 (GH30-7) and 8 (GH30-8). Unlike for the well-studied GH30-8 enzymes, the structural and functional characteristics of GH30-7 enzymes remain poorly understood. Here, we report the catalytic properties and three-dimensional structure of GH30-7 xylanase B (Xyn30B) identified from the cellulolytic fungus Talaromyces cellulolyticus Xyn30B efficiently degraded glucuronoxylan to acidic xylooligosaccharides (XOSs), including an α-1,2-linked 4- O -methyl-d-glucuronosyl substituent (MeGlcA). Rapid analysis with negative-mode electrospray-ionization multistage MS (ESI(-)-MS n ) revealed that the structures of the acidic XOS products are the same as those of the hydrolysates (MeGlcA 2 Xyl n , n > 2) obtained with typical glucuronoxylanases. Acidic XOS products were further degraded by Xyn30B, releasing first xylobiose and then xylotetraose and xylohexaose as transglycosylation products. This hydrolase reaction was unique to Xyn30B, and the substrate was cleaved at the xylobiose unit from its nonreducing end, indicating that Xyn30B is a bifunctional enzyme possessing both endo-glucuronoxylanase and exo-xylobiohydrolase activities. The crystal structure of Xyn30B was determined as the first structure of a GH30-7 xylanase at 2.25 Å resolution, revealing that Xyn30B is composed of a pseudo-(α/β) 8 -catalytic domain, lacking an α6 helix, and a small β-rich domain. This structure and site-directed mutagenesis clarified that Arg 46 , conserved in GH30-7 glucuronoxylanases, is a critical residue for MeGlcA appendage-dependent xylan degradation. The structural comparison between Xyn30B and the GH30-8 enzymes suggests that Asn 93 in the β2-α2 loop is involved in xylobiohydrolase activity. In summary, our findings indicate that Xyn30B is a bifunctional endo- and exo-xylanase.
Organizational Affiliation:
From the Bioconversion Group and.