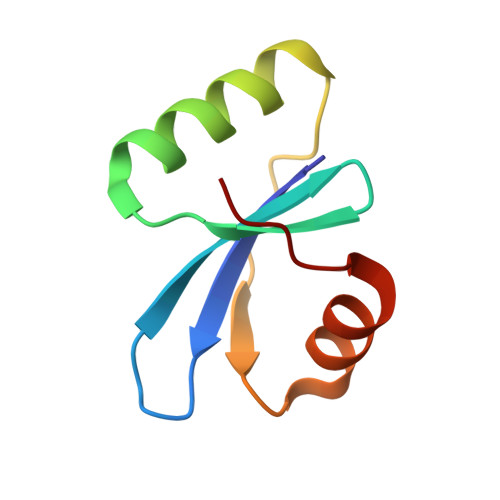
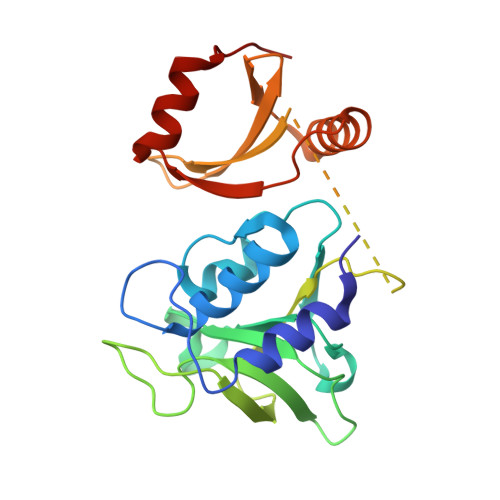
Structural and functional insights into a novel two-component endolysin encoded by a single gene in Enterococcus faecalis phage.
Zhou, B., Zhen, X., Zhou, H., Zhao, F., Fan, C., Perculija, V., Tong, Y., Mi, Z., Ouyang, S.(2020) PLoS Pathog 16: e1008394-e1008394
- PubMed: 32176738
- DOI: https://doi.org/10.1371/journal.ppat.1008394
- Primary Citation of Related Structures:
6IST, 6L00 - PubMed Abstract:
Using bacteriophage-derived endolysins as an alternative strategy for fighting drug-resistant bacteria has recently been garnering renewed interest. However, their application is still hindered by their narrow spectra of activity. In our previous work, we demonstrated that the endolysin LysIME-EF1 possesses efficient bactericidal activity against multiple strains of Enterococcus faecalis (E. faecalis). Herein, we observed an 8 kDa fragment and hypothesized that it contributes to LysIME-EF1 lytic activity. To examine our hypothesis, we determined the structure of LysIME-EF1 at 1.75 Å resolution. LysIME-EF1 exhibits a unique architecture in which one full-length LysIME-EF1 forms a tetramer with three additional C-terminal cell-wall binding domains (CBDs) that correspond to the abovementioned 8 kDa fragment. Furthermore, we identified an internal ribosomal binding site (RBS) and alternative start codon within LysIME-EF1 gene, which are demonstrated to be responsible for the translation of the truncated CBD. To elucidate the molecular mechanism for the lytic activity of LysIME-EF1, we combined mutagenesis, lytic activity assays and in vivo animal infection experiments. The results confirmed that the additional LysIME-EF1 CBDs are important for LysIME-EF1 architecture and its lytic activity. To our knowledge, this is the first determined structure of multimeric endolysin encoded by a single gene in E. faecalis phages. As such, it may provide valuable insights into designing potent endolysins against the opportunistic pathogen E. faecalis.
Organizational Affiliation:
The Key Laboratory of Innate Immune Biology of Fujian Province, Provincial University Key Laboratory of Cellular Stress Response and Metabolic Regulation, Biomedical Research Center of South China, Key Laboratory of OptoElectronic Science and Technology for Medicine of the Ministry of Education, College of Life Sciences, Fujian Normal University, Fuzhou, China.