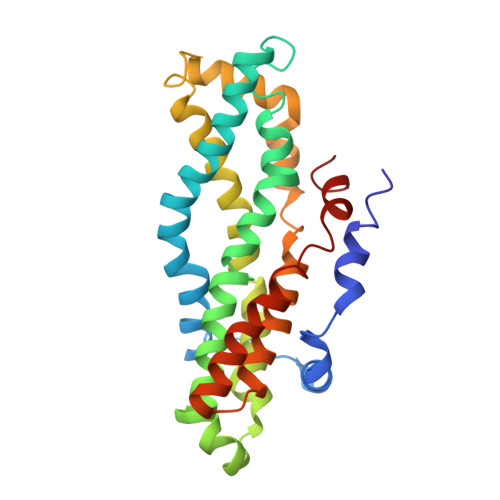
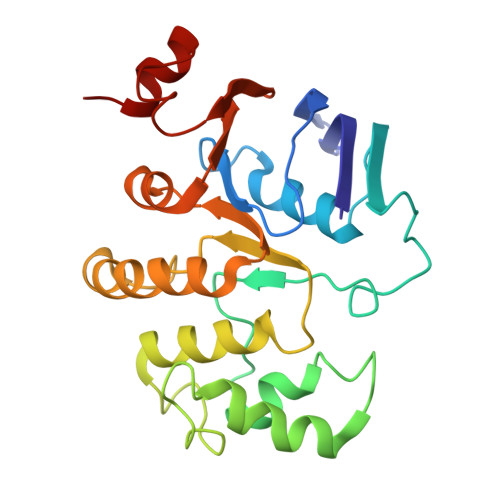
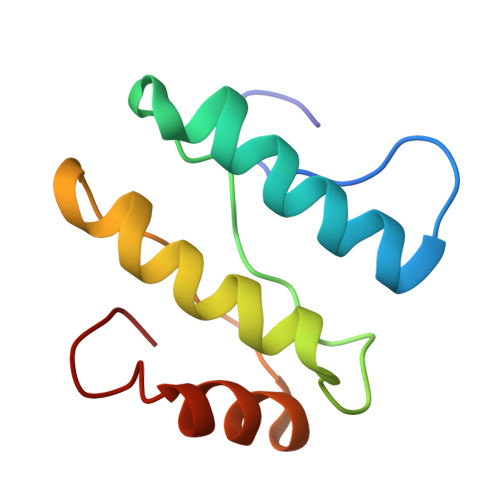
The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane.
Kamischke, C., Fan, J., Bergeron, J., Kulasekara, H.D., Dalebroux, Z.D., Burrell, A., Kollman, J.M., Miller, S.I.(2019) Elife 8
- PubMed: 30638443
- DOI: https://doi.org/10.7554/eLife.40171
- Primary Citation of Related Structures:
6IC4 - PubMed Abstract:
The outer membrane (OM) of Gram-negative bacteria serves as a selective permeability barrier that allows entry of essential nutrients while excluding toxic compounds, including antibiotics. The OM is asymmetric and contains an outer leaflet of lipopolysaccharides (LPS) or lipooligosaccharides (LOS) and an inner leaflet of glycerophospholipids (GPL). We screened Acinetobacter baumannii transposon mutants and identified a number of mutants with OM defects, including an ABC transporter system homologous to the Mla system in E. coli. We further show that this opportunistic, antibiotic-resistant pathogen uses this multicomponent protein complex and ATP hydrolysis at the inner membrane to promote GPL export to the OM. The broad conservation of the Mla system in Gram-negative bacteria suggests the system may play a conserved role in OM biogenesis. The importance of the Mla system to Acinetobacter baumannii OM integrity and antibiotic sensitivity suggests that its components may serve as new antimicrobial therapeutic targets.
Organizational Affiliation:
Department of Microbiology, University of Washington, Seattle, United States.