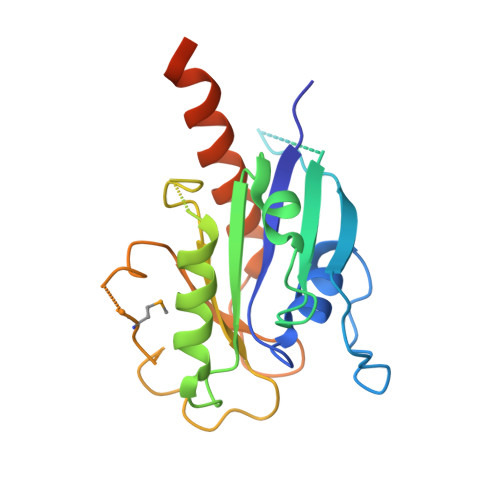
Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly.
Wachter, S., Jung, J., Shafiq, S., Basquin, J., Fort, C., Bastin, P., Lorentzen, E.(2019) EMBO J 38
- PubMed: 30940671
- DOI: https://doi.org/10.15252/embj.2018101251
- Primary Citation of Related Structures:
6IA7, 6IAE, 6IAN - PubMed Abstract:
Intraflagellar transport (IFT) relies on motor proteins and the IFT complex to construct cilia and flagella. The IFT complex subunit IFT22/RabL5 has sequence similarity with small GTPases although the nucleotide specificity is unclear because of non-conserved G4/G5 motifs. We show that IFT22 specifically associates with G-nucleotides and present crystal structures of IFT22 in complex with GDP, GTP, and with IFT74/81. Our structural analysis unravels an unusual GTP/GDP-binding mode of IFT22 bypassing the classical G4 motif. The GTPase switch regions of IFT22 become ordered upon complex formation with IFT74/81 and mediate most of the IFT22-74/81 interactions. Structure-based mutagenesis reveals that association of IFT22 with the IFT complex is essential for flagellum construction in Trypanosoma brucei although IFT22 GTP-loading is not strictly required.
Organizational Affiliation:
Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Martinsried, Germany.