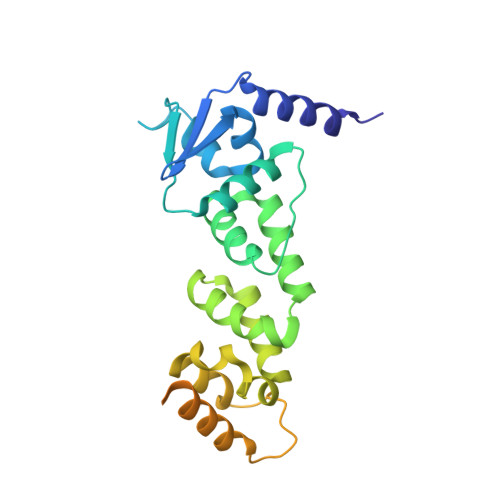
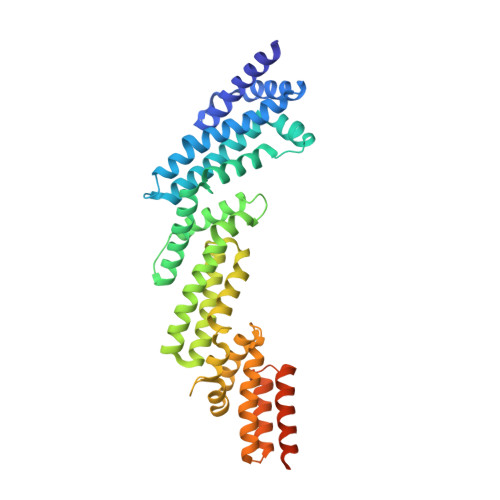
Molecular basis of cullin-3 (Cul3) ubiquitin ligase subversion by vaccinia virus protein A55.
Gao, C., Pallett, M.A., Croll, T.I., Smith, G.L., Graham, S.C.(2019) J Biol Chem 294: 6416-6429
- PubMed: 30819806
- DOI: https://doi.org/10.1074/jbc.RA118.006561
- Primary Citation of Related Structures:
6I2M - PubMed Abstract:
BTB-Kelch proteins are substrate-specific adaptors for cullin-3 (Cul3) RING-box-based E3 ubiquitin ligases, mediating protein ubiquitylation for subsequent proteasomal degradation. Vaccinia virus encodes three BTB-Kelch proteins: A55, C2, and F3. Viruses lacking A55 or C2 have altered cytopathic effects in cultured cells and altered pathology in vivo Previous studies have shown that the ectromelia virus orthologue of A55 interacts with Cul3 in cells. We report that the N-terminal BTB-BACK (BB) domain of A55 binds directly to the Cul3 N-terminal domain (Cul3-NTD), forming a 2:2 complex in solution. We solved the structure of an A55BB/Cul3-NTD complex from anisotropic crystals diffracting to 2.3/3.7 Å resolution in the best/worst direction, revealing that the overall interaction and binding interface closely resemble the structures of cellular BTB/Cul3-NTD complexes, despite low sequence identity between A55 and cellular BTB domains. Surprisingly, despite this structural similarity, the affinity of Cul3-NTD for A55BB was stronger than for cellular BTB proteins. Glutamate substitution of the A55 residue Ile-48, adjacent to the canonical φ X (D/E) Cul3-binding motif, reduced affinity of A55BB for Cul3-NTD by at least 2 orders of magnitude. Moreover, Ile-48 and the φ X (D/E) motif are conserved in A55 orthologues from other poxviruses, but not in the vaccinia virus proteins C2 or F3. The high-affinity interaction between A55BB and Cul3-NTD suggests that, in addition to directing the Cul3-RING E3 ligase complex to degrade cellular/viral target proteins that are normally unaffected, A55 may also sequester Cul3 from cellular adaptor proteins, thereby protecting substrates of these cellular adaptors from ubiquitylation and degradation.
Organizational Affiliation:
From the Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge CB2 1QP and.