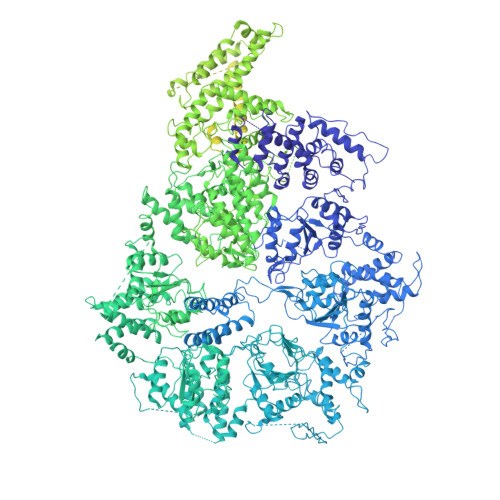
The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Sosnowski, P., Urnavicius, L., Boland, A., Fagiewicz, R., Busselez, J., Papai, G., Schmidt, H.(2018) Elife 7
- PubMed: 30460895
- DOI: https://doi.org/10.7554/eLife.39163
- Primary Citation of Related Structures:
6HYD, 6HYP, 6I26, 6I27 - PubMed Abstract:
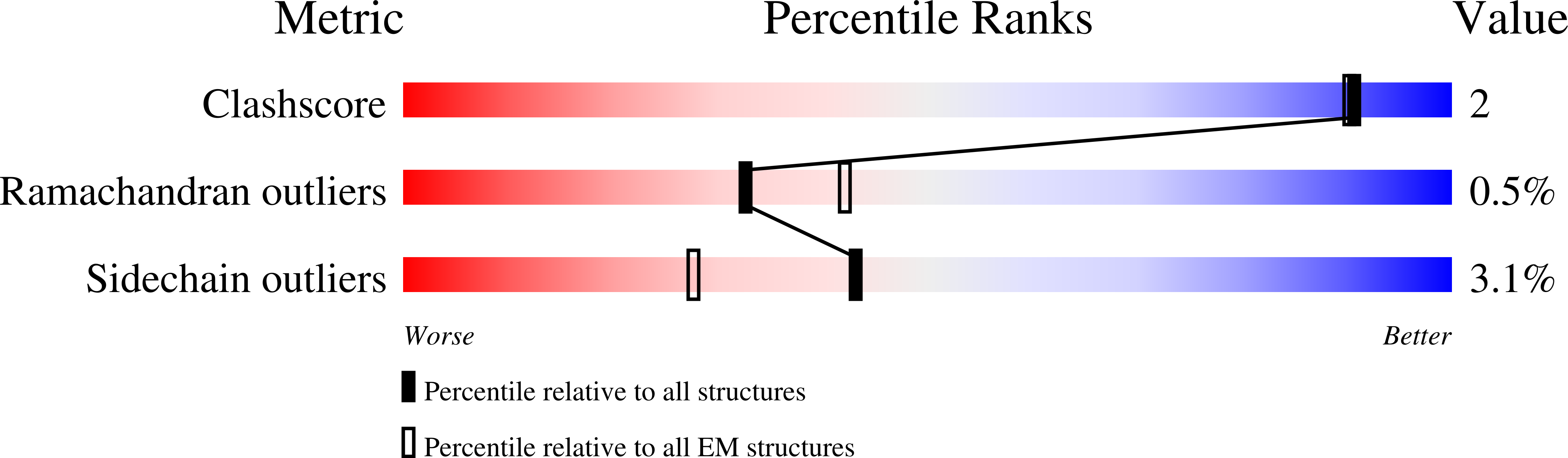
The biogenesis of 60S ribosomal subunits is initiated in the nucleus where rRNAs and proteins form pre-60S particles. These pre-60S particles mature by transiently interacting with various assembly factors. The ~5000 amino-acid AAA+ ATPase Rea1 (or Midasin) generates force to mechanically remove assembly factors from pre-60S particles, which promotes their export to the cytosol. Here we present three Rea1 cryoEM structures. We visualise the Rea1 engine, a hexameric ring of AAA+ domains, and identify an α-helical bundle of AAA2 as a major ATPase activity regulator. The α-helical bundle interferes with nucleotide-induced conformational changes that create a docking site for the substrate binding MIDAS domain on the AAA +ring. Furthermore, we reveal the architecture of the Rea1 linker, which is involved in force generation and extends from the AAA+ ring. The data presented here provide insights into the mechanism of one of the most complex ribosome maturation factors.
Organizational Affiliation:
Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France.