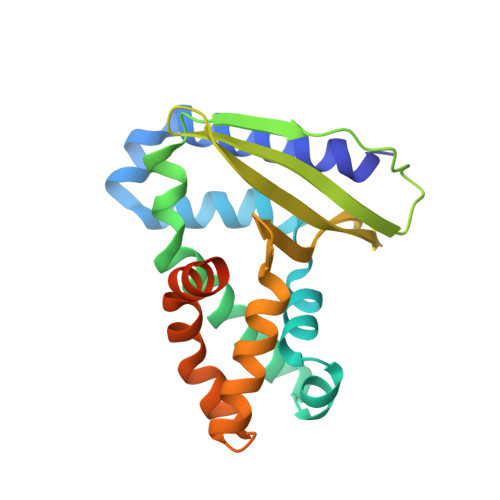
The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm.
Yero, D., Diaz-Lobo, M., Costenaro, L., Conchillo-Sole, O., Mayo, A., Ferrer-Navarro, M., Vilaseca, M., Gibert, I., Daura, X.(2021) Commun Biol 4: 448-448
- PubMed: 33837253
- DOI: https://doi.org/10.1038/s42003-021-01968-8
- Primary Citation of Related Structures:
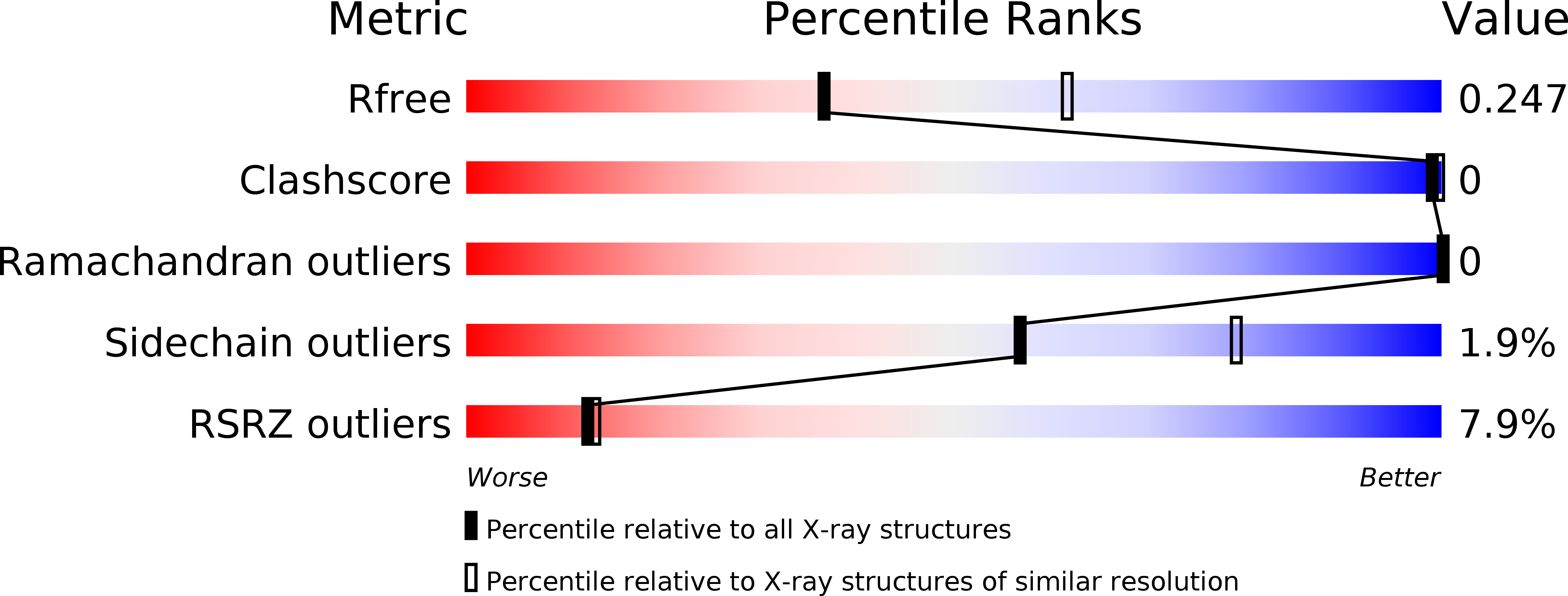
6HSY - PubMed Abstract:
In Pseudomonas aeruginosa, Ttg2D is the soluble periplasmic phospholipid-binding component of an ABC transport system thought to be involved in maintaining the asymmetry of the outer membrane. Here we use the crystallographic structure of Ttg2D at 2.5 Å resolution to reveal that this protein can accommodate four acyl chains. Analysis of the available structures of Ttg2D orthologs shows that they conform a new substrate-binding-protein structural cluster. Native and denaturing mass spectrometry experiments confirm that Ttg2D, produced both heterologously and homologously and isolated from the periplasm, can carry two diacyl glycerophospholipids as well as one cardiolipin. Binding is notably promiscuous, allowing the transport of various molecular species. In vitro binding assays coupled to native mass spectrometry show that binding of cardiolipin is spontaneous. Gene knockout experiments in P. aeruginosa multidrug-resistant strains reveal that the Ttg2 system is involved in low-level intrinsic resistance against certain antibiotics that use a lipid-mediated pathway to permeate through membranes.
Organizational Affiliation:
Institut de Biotecnologia i de Biomedicina (IBB), Universitat Autònoma de Barcelona (UAB), Barcelona, Spain.