In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex.
Rapisarda, C., Cherrak, Y., Kooger, R., Schmidt, V., Pellarin, R., Logger, L., Cascales, E., Pilhofer, M., Durand, E., Fronzes, R.(2019) EMBO J
- PubMed: 30877094
- DOI: https://doi.org/10.15252/embj.2018100886
- Primary Citation of Related Structures:
6HS7 - PubMed Abstract:
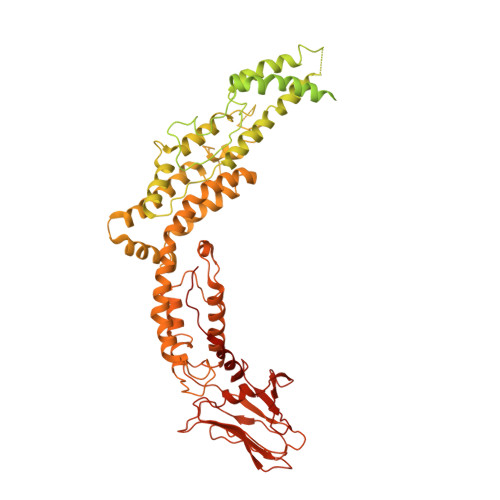
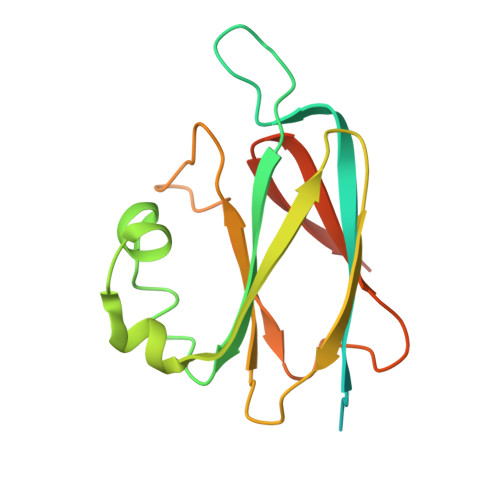
Bacteria have evolved macromolecular machineries that secrete effectors and toxins to survive and thrive in diverse environments. The type VI secretion system (T6SS) is a contractile machine that is related to Myoviridae phages. It is composed of a phage tail-like structure inserted in the bacterial cell envelope by a membrane complex (MC) comprising the TssJ, TssL and TssM proteins. We previously reported the low-resolution negative-stain electron microscopy structure of the enteroaggregative Escherichia coli MC and proposed a rotational 5-fold symmetry with a TssJ:TssL:TssM stoichiometry of 2:2:2. Here, cryo-electron tomography analyses of the T6SS MC confirm the 5-fold symmetry in situ and identify the regions of the structure that insert into the bacterial membranes. A high-resolution model obtained by single-particle cryo-electron microscopy highlights new features: five additional copies of TssJ, yielding a TssJ:TssL:TssM stoichiometry of 3:2:2, an 11-residue loop in TssM, protruding inside the lumen of the MC and constituting a functionally important periplasmic gate, and hinge regions. Based on these data, we propose an updated model on MC structure and dynamics during T6SS assembly and function.
Organizational Affiliation:
CNRS UMR 5234 Microbiologie Fondamentale et Pathogénicité, Bordeaux, France.