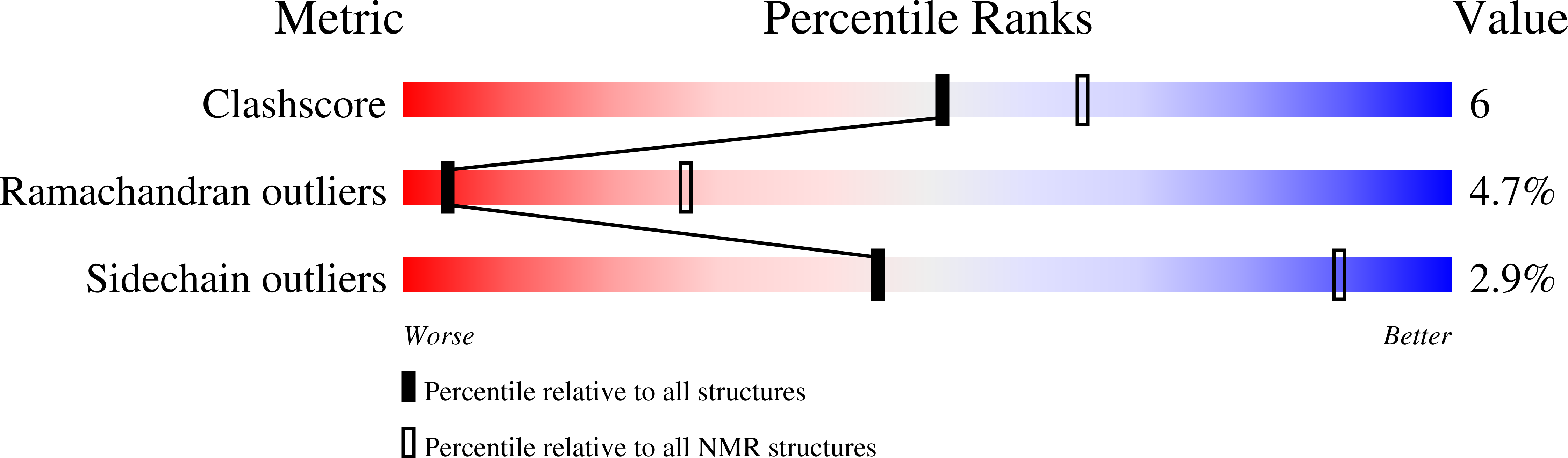Secreted amyloid-beta precursor protein functions as a GABABR1a ligand to modulate synaptic transmission.
Rice, H.C., de Malmazet, D., Schreurs, A., Frere, S., Van Molle, I., Volkov, A.N., Creemers, E., Vertkin, I., Nys, J., Ranaivoson, F.M., Comoletti, D., Savas, J.N., Remaut, H., Balschun, D., Wierda, K.D., Slutsky, I., Farrow, K., De Strooper, B., de Wit, J.(2019) Science 363
- PubMed: 30630900
- DOI: https://doi.org/10.1126/science.aao4827
- Primary Citation of Related Structures:
6HKC - PubMed Abstract:
Amyloid-β precursor protein (APP) is central to the pathogenesis of Alzheimer's disease, yet its physiological function remains unresolved. Accumulating evidence suggests that APP has a synaptic function mediated by an unidentified receptor for secreted APP (sAPP). Here we show that the sAPP extension domain directly bound the sushi 1 domain specific to the γ-aminobutyric acid type B receptor subunit 1a (GABA B R1a). sAPP-GABA B R1a binding suppressed synaptic transmission and enhanced short-term facilitation in mouse hippocampal synapses via inhibition of synaptic vesicle release. A 17-amino acid peptide corresponding to the GABA B R1a binding region within APP suppressed in vivo spontaneous neuronal activity in the hippocampus of anesthetized Thy1-GCaMP6s mice. Our findings identify GABA B R1a as a synaptic receptor for sAPP and reveal a physiological role for sAPP in regulating GABA B R1a function to modulate synaptic transmission.
Organizational Affiliation:
VIB Center for Brain & Disease Research, Leuven, Belgium.