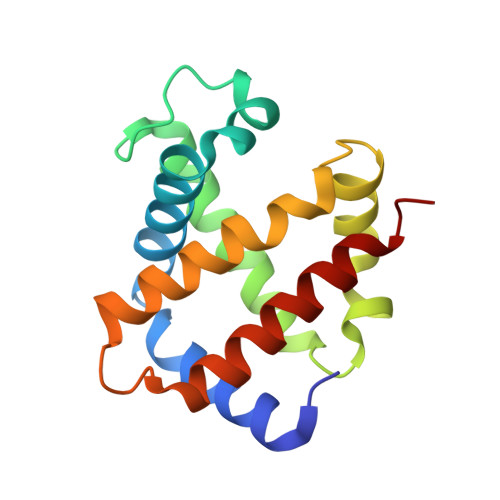
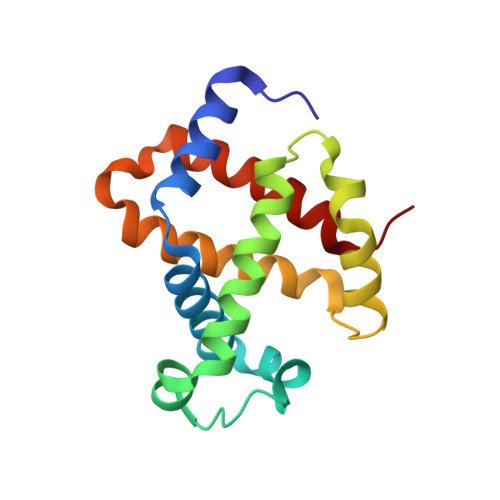
The crystal structure of haemoglobin from Atlantic cod.
Helland, R., Bjorkeng, E.K., Rothweiler, U., Sydnes, M.O., Pampanin, D.M.(2019) Acta Crystallogr F Struct Biol Commun 75: 537-542
- PubMed: 31397324
- DOI: https://doi.org/10.1107/S2053230X1900904X
- Primary Citation of Related Structures:
6HIT - PubMed Abstract:
The crystal structure of haemoglobin from Atlantic cod has been solved to 2.54 Å resolution. The structure consists of two tetramers in the crystallographic asymmetric unit. The structure of haemoglobin obtained from one individual cod suggests polymorphism in the tetrameric assembly.
Organizational Affiliation:
NorStruct, Department of Chemistry, Faculty of Science and Technology, UiT - The Arctic University of Norway, NO-9037 Tromsø, Norway.