A Fragment-Derived Clinical Candidate for Antagonism of X-Linked and Cellular Inhibitor of Apoptosis Proteins: 1-(6-[(4-Fluorophenyl)methyl]-5-(hydroxymethyl)-3,3-dimethyl-1 H,2 H,3 H-pyrrolo[3,2- b]pyridin-1-yl)-2-[(2 R,5 R)-5-methyl-2-([(3R)-3-methylmorpholin-4-yl]methyl)piperazin-1-yl]ethan-1-one (ASTX660).
Johnson, C.N., Ahn, J.S., Buck, I.M., Chiarparin, E., Day, J.E.H., Hopkins, A., Howard, S., Lewis, E.J., Martins, V., Millemaggi, A., Munck, J.M., Page, L.W., Peakman, T., Reader, M., Rich, S.J., Saxty, G., Smyth, T., Thompson, N.T., Ward, G.A., Williams, P.A., Wilsher, N.E., Chessari, G.(2018) J Med Chem 61: 7314-7329
- PubMed: 30091600
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00900
- Primary Citation of Related Structures:
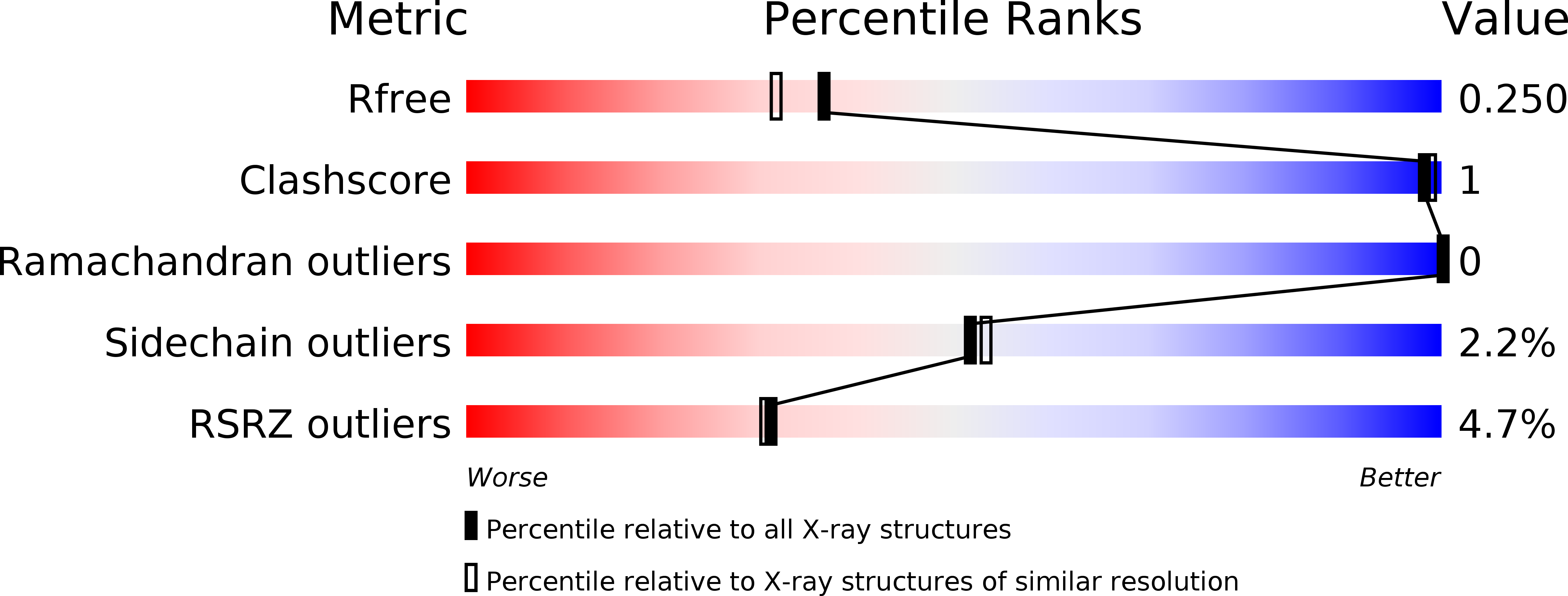
6H6Q, 6H6R - PubMed Abstract:
Inhibitor of apoptosis proteins (IAPs) are promising anticancer targets, given their roles in the evasion of apoptosis. Several peptidomimetic IAP antagonists, with inherent selectivity for cellular IAP (cIAP) over X-linked IAP (XIAP), have been tested in the clinic. A fragment screening approach followed by structure-based optimization has previously been reported that resulted in a low-nanomolar cIAP1 and XIAP antagonist lead molecule with a more balanced cIAP-XIAP profile. We now report the further structure-guided optimization of the lead, with a view to improving the metabolic stability and cardiac safety profile, to give the nonpeptidomimetic antagonist clinical candidate 27 (ASTX660), currently being tested in a phase 1/2 clinical trial (NCT02503423).
Organizational Affiliation:
Astex Pharmaceuticals , 436 Cambridge Science Park, Milton Road , Cambridge CB4 0QA , United Kingdom.